Double-stranded RNA Recognition in Gene Silencing Pathway
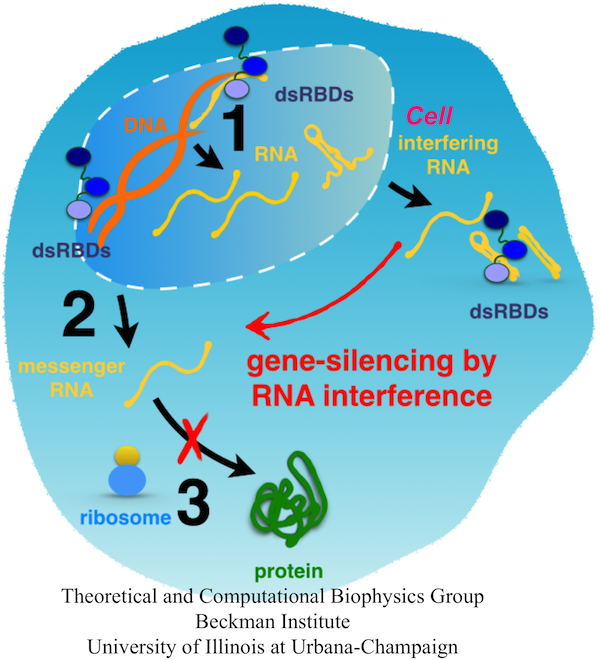
Fig. 1. A scheme of the flow of genetic information inside a cell. DNA is transcribed into mRNA or non-coding RNA (step 1). RNA is processed and can remain in the nucleus or be exported into the cytoplasm (step 2). mRNA is read by ribosomes that synthesize the respective proteins (step 3); protein synthesis can be blocked by interfering RNA.
Function and development of every organism is encoded in DNA of its genome. Within the genome, genes store codes of all our proteins, main actors of our cells. While genes comprise less than 2% of our DNA, up to 80% of the genome is copied into RNA molecules that can execute the instructions stored in DNA. For example, genes in their RNA forms serve as templates for synthesis of new proteins, whereas non-coding RNA can bind the copied genes and silence protein synthesis. Frequently, gene-silencing RNA is activated with the help of conserved double stranded RNA binding domains (dsRBDs), which recognize its characteristic duplex form. For example, a Dicer enzyme, which contains one dsRBD, binds to and slices dsRNA into 21-22 nucleotide long pieces that silence mRNA, and transactivation response RNA binding protein (TRBP) binds dsRNA and helps to position dsRNA in proper conformation for its cuting by Dicer.
Interestingly, TRBP does not bind to duplex types other than dsRNA, namely dsDNA and DNA-RNA hybrids. Many other dsRBD proteins specifically bind to dsRNA duplexes, whereas some also weakly bind to DNA-RNA hybrid duplexes. Considering the experimental data about dsRBD binding targets, there are two questions of particular interest: (1) How do dsRBDs discriminate between nucleic acid duplexes of similar structures, such as dsRNA, DNA-RNA, and dsDNA? (2) How do dsRBDs discriminate between dsRNA of different sequences, abundant in the cell nuclei and cytoplasm? We address the first question in our study, choosing a representative dsRBD, the second dsRBD of TRBP (TRBP-RBD2).
dsRBD binding to dsRNA
To examine how TRBP-RBD2 (and in general, dsRBDs) recognize dsRNA, we performed MD simulations of the TRBP-RBD2:dsRNA complex, shown in Fig. 1 (a). TRBP- RBD2 binds to dsRNA along the duplex axis and across three grooves of the duplex (minor- major-minor grooves), similarly to other dsRBDs. dsRNA minor grooves are lined with 2'-hydroxyl groups on sugar rings of RNA nucleotides, and the major grooves are lined with the negatively charged phosphate groups. dsRNA duplex assumes the A-form, in which the widths of minor and major grooves are comparable, as marked in Fig. 1 (a). In TRBP-RBD2:dsRNA complex, there are numerous contacts between TRBP-RBD2 and minor and major grooves of dsRNA.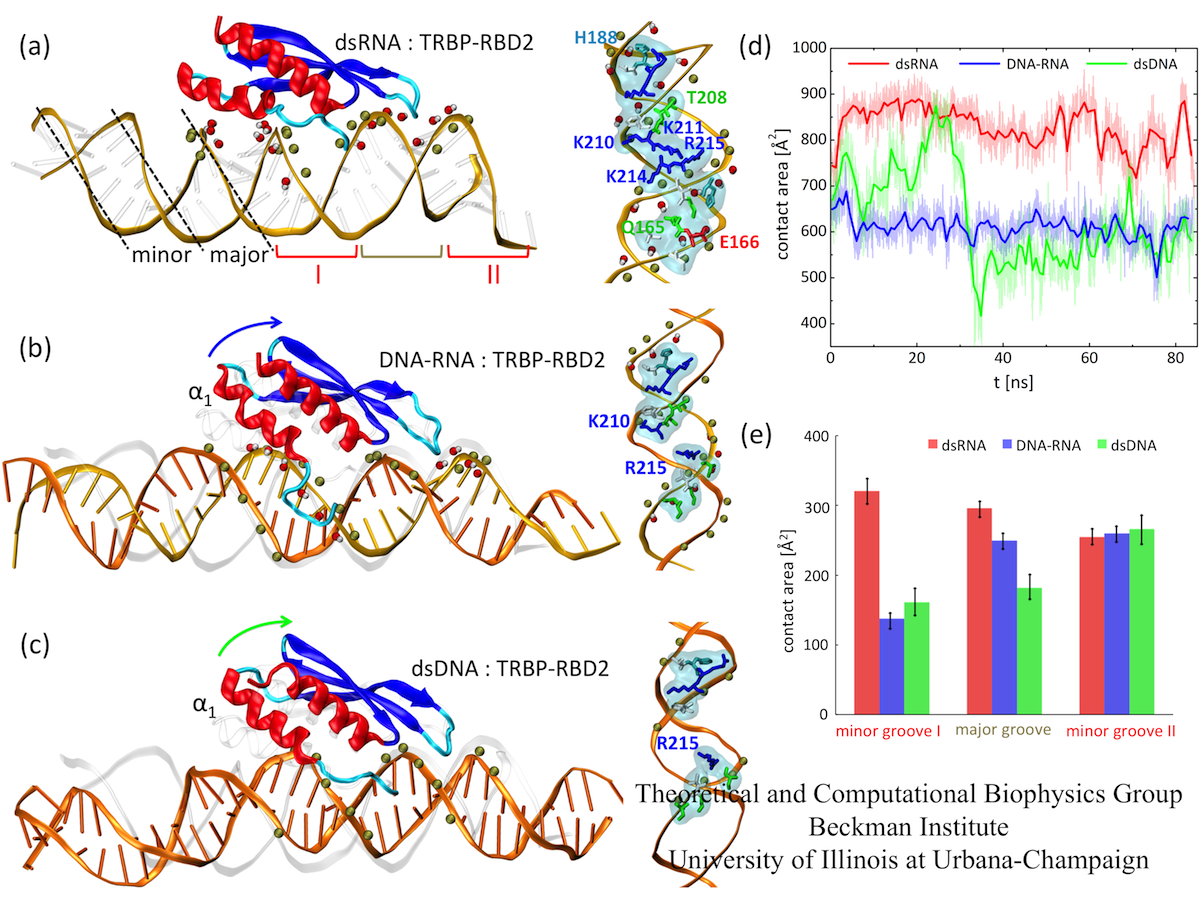
Fig. 2. Binding of TRBP-RBD2 to dsRNA (a), DNA-RNA (b), and dsDNA (c). (d) Contact areas between TRBP-RBD2 and the three duplexes. (e) Contact areas between TRBP-RBD2 and individual duplex grooves.
TRBP-RBD2 discriminates between dsRNA and DNA-containing duplexes
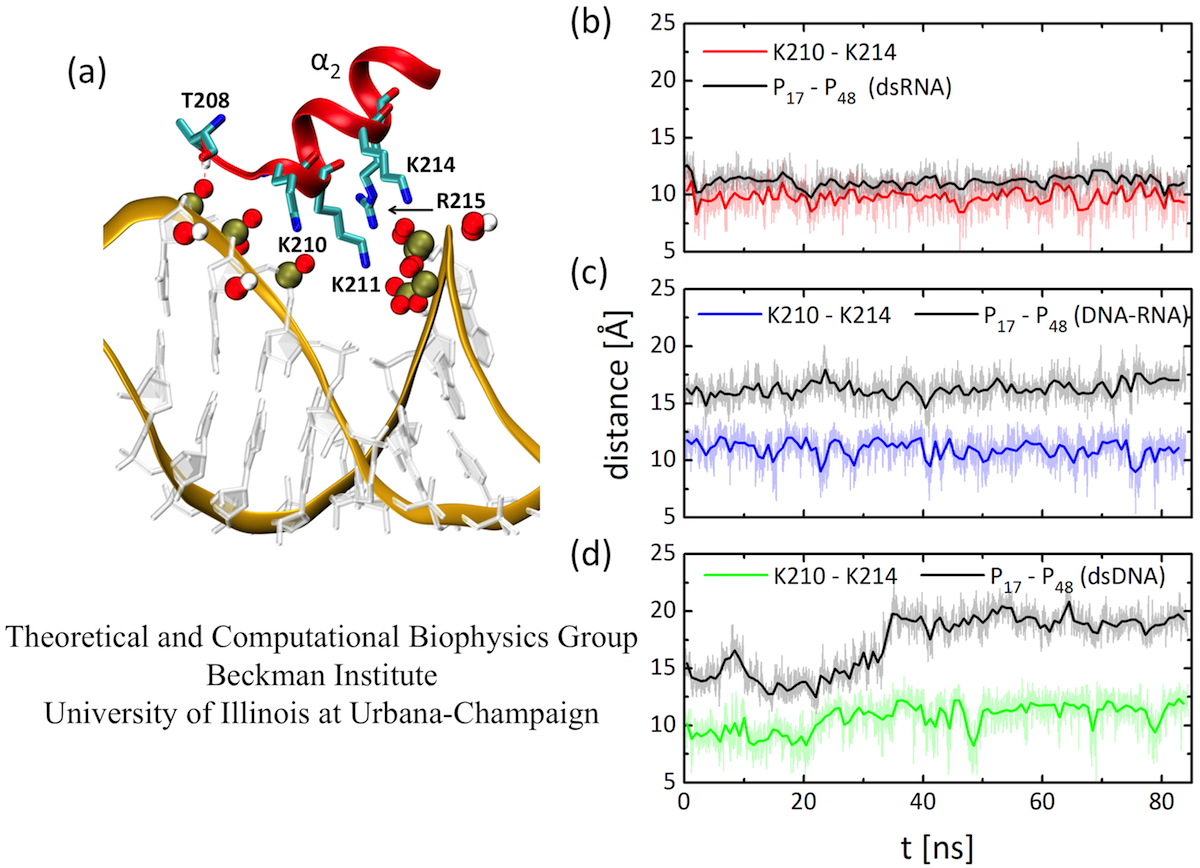
Fig. 3. (a) Binding mode of TRBP-RBD2 at the dsRNA major groove. (b-d) Distances between Lys 210 and Lys 214 residues for TRBP-RBD2 bound to three studied duplexes, compared to major groove widths of the three duplexes. The Lys 210 - Lys 214 pair fits well only into the major groove of dsRNA.
To examine how TRBP-RBD2 discriminates between dsRNA and DNA-containing duplexes, we simulated TRBP-RBD2 initially binding to nucleic acid duplexes containing DNA. Snapshots of TRBP-RBD2 in complex with DNA-RNA and dsDNA, captured at the end of 100 ns long simulations, are shown in Fig.1(b) and (c), respectively. DNA-RNA and dsDNA duplexes stay in contact with a smaller number of TRBP-RBD2 residues than does dsRNA. There is loss of TRBP- RBD2 binding to minor groove I (helix α1 lifts from the groove)) and the major groove. However, the numbers of TRBP-RBD2 contacts to the minor groove II are very similar for all three studied duplexes. The contact areas of TRBP-RBD2 and the three duplexes, shown in Fig. 1 (d-e), have been evaluated during the last 85 ns of the simulations. The contact area decreases as the DNA content of the duplex increases, especially in minor groove I and the major groove, confirming that TRBP-RBD2 binds most favorably to its cognate substrate, dsRNA, in agreement with experimental observations. The results indicate that TRBP-RBD2 recognizes dsRNA on the basis of interactions in minor groove I and in the major groove.
A particularly important region for dsRBD:dsRNA binding is the major groove. The binding mode of TRBP-RBD2 to the major groove of dsRNA is shown in Figure 3. Five amino acid residues make direct contact with the dsRNA backbone at the major groove. A close inspection of the binding mode shows that Lys210 and Lys214 could act as a ruler of the dsRNA major groove width, since they bridge phosphate groups on two sides of the dsRNA major groove. Therefore, we examine how Lys210-Lys214 distance compares to major groove groove widths of the three duplexes. The results show evidence for a "register" fit of Lys210-Lys214, as the lysine pair fits exactly above the phosphate groups that coordinate them. The distance between positively charged tips of Lys210 and Lys214 residues is always in the range ~9-13 Å, whereas the distance between the bridging phosphate groups increases from ~11 Å for dsRNA to ~17 Å for DNA-RNA and to ~20 Å for dsDNA. The Lys210- Lys214 pair fits poorly in the major grooves of DNA-RNA and dsDNA duplexes because the major grooves of these duplexes are significantly wider, and the pair cannot readjust its width.
Duplex form plays an important role in duplex recognition
Next, we examine the form of the studied duplexes. Several structural features can be clearly discerned in Fig. 4. First, the major groove is narrowest for dsRNA, wider for DNA-RNA, and widest for dsDNA. Second, minor grooves I of DNA-RNA and dsDNA can be almost perfectly superimposed on each other, but not to the more compressed minor groove I of dsRNA; only dsRNA achieves stable binding to TRBP-RBD2. Third, the minor groove II of DNA-RNA assumes a form almost identical to that of dsRNA (also seen in Fig. 4(d), in the groove marked by the black arrow), whereas dsDNA exhibits a much narrower minor groove II. Therefore, through the presence of TRBP-RBD2, the minor grooves of DNA-RNA adopt either dsRNA or dsDNA forms, depending on the interactions with the protein. The observed B- to A-form change of DNA nucleotides and the resulting minor groove width modulation potentially explain how other dsRBD proteins, such as 4F can also bind to DNA-RNA hybrids.
We quantify the duplex form by comparing helical rise of the simulated duplexes. Fig. 4 shows that helical rise of perfect A-form dsRNA is ~2.5 Å, whereas for B-form dsDNA it is 3.4 Å. The results show for dsRNA a perfect A-form, except in one region of widened major groove. For DNA-RNA, the helical rise is between A- and B-forms, as observed in other studies of free DNA-RNA duplexes; for dsDNA the helical rise is again between A- and B-form, but very close to the B-form value. dsRNA has the smallest helical rise and remains closest to the A-form, which is recognized by TRBP-RBD2.
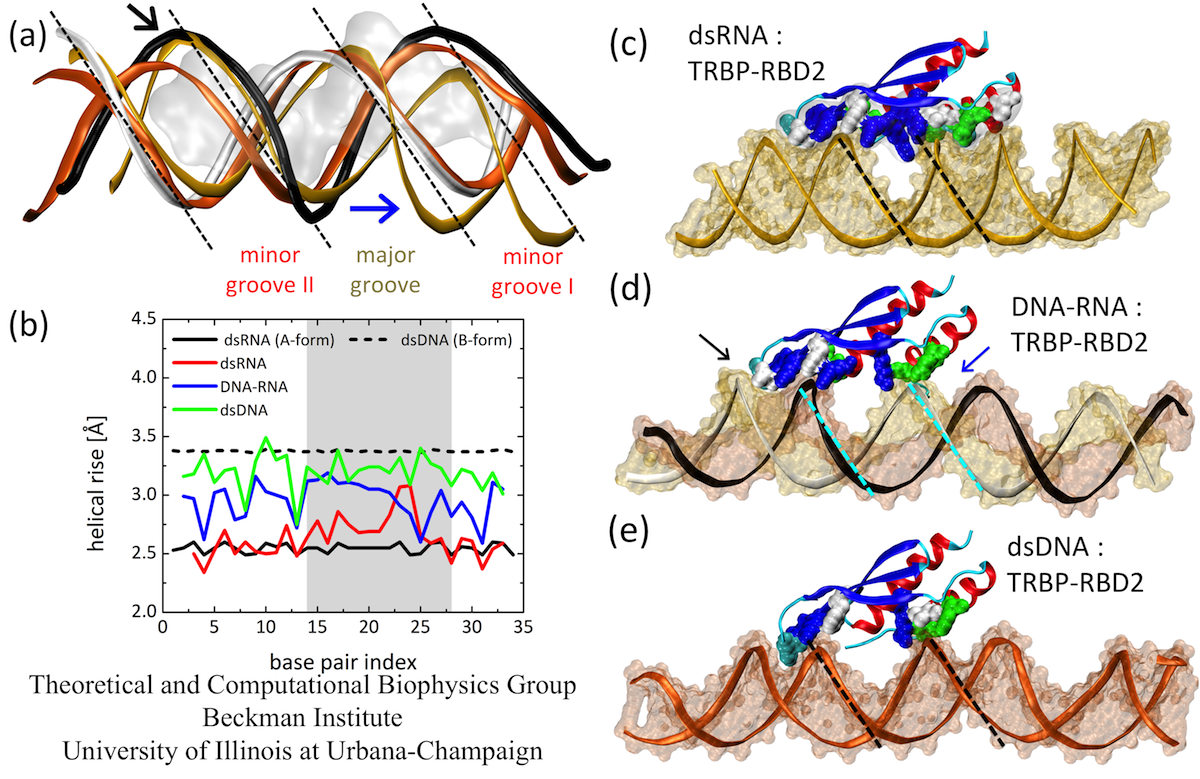
Fig. 4. (a) Duplex forms at TRBP-RBD2 binding sites. (b) Helical rise for duplex base pairs of dsRNA, DNA-RNA and dsDNA. (c-e) Average structures of three complexes have marked differences in forms (groove widths and depths). Minor grooves of DNA-RNA can assume B-form (deep and narrow, marked by blue arrow) or A-form (wide and shallow, marked by black arrow).
TCBG Publications
Substrate recognition and specificity of double-stranded RNA binding proteins. Lela Vukovic, Hye Ran Koh, Sua Myong, and Klaus Schulten. Biochemistry, 53:3457-3466, 2014.
Investigators
- Lela Vukovic
- Hye Ran Koh
- Sua Myong
- Klaus Schulten
maintained by Lela Vukovic



