Bacteriorhodopsin
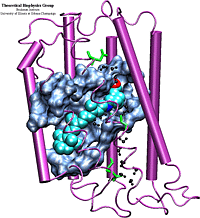
Bacteriorhodopsin is a trans-membrane protein found in the cellular membrane of Halobacterium salinarium, which functions as a light-driven proton pump. With the recent determination of the structure of bR by electron cryo-microscopy [1], simulation of the proton pump cycle has become feasible. Previous refinements of the bR structure and simulations of the bR proton pump cycle [2,3,4] in vacuo may account for many experimental observations. However, some controversial issues still remain, which presumably can be resolved by simulating the protein in its native membrane environment. Utilization of new faster processors will allow the group to carry out the following important tests and simulations on the protein inside a membrane have not been possible, until now, due to computer time restrictions.
- (1) Include bR in its natural membrane environment (see the bR and purple
membrane web page).
- (2) Monitor the effect that insertion of bR has on the membrane; specifically, it will be observed if nearby lipids change their conformational and dynamical behavior significantly, or if they remain relatively unaffected.
- (3) After equilibration and examination of the protein-membrane complex, simulate the early stages of the bR photocycle.
- (4) Compare the results of these ``full system'' molecular dynamics simulations with the ones carried out for bacteriorhodopsin in vacuum which will allow one to draw conclusions about the general accuracy of vacuum simulations.
- (2) Monitor the effect that insertion of bR has on the membrane; specifically, it will be observed if nearby lipids change their conformational and dynamical behavior significantly, or if they remain relatively unaffected.
Some important stages of the bR photocycle are poorly reproduced by conventional molecular dynamics simulations due to the empirical nature of the force field employed in this method. This deficiency is particularly noticeable when processes such as photoisomerization or proton transfer are simulated. These processes require quantum mechanical evaluation of forces, since the latter are not covered by the semiempirical force field available. Due to lack of detailed quantum mechanical calculations of the retinal excited state, a particular mechanism of the initial photoisomerization step in the bR proton pump cycle still remains as an issue of controversy [5]. Some preliminary ab initio quantum mechanical calculations of the proton transfer process have been computed on model systems [6]; however, no attempt has been made to simulate this process in the real protein environment within the bacteriorhodopsin active site.
It is proposed to utilize a combination of ab initio quantum mechanical and molecular mechanical approaches to model (1) the initial retinal photoisomerization and (2) the proton transfer stages in the bacteriorhodopsin photocycle. A number of research efforts have combined quantum mechanics programs with molecular mechanics programs [7,8,9]. Although such implementations are straightforward, dynamical ab initio calculations were considered to be computationally prohibitive. In modelling the bacteriorhodopsin photocycle, a combination of the ab initio quantum mechanical and molecular mechanical methods will be used for the first time to do direct dynamical simulations. Such a unique project is now feasible because (1) the time scales of the processes of interest (photoisomerization and proton transfer in bR) are very short, ranging from tens to hundreds of femtoseconds [10]; (2) the number of heavy atoms to be simulated with quantum mechanics is moderate, varying from ten to twenty atoms for different calculations; (3) the availability of new high performance workstations has put the computational requirements for such ambitious calculations within reach.
More information on the dynamics of bacteriorhodopsin and of the purple membrane.
 Emad Tajkhorshid - MD
simulations, QM calculations
Emad Tajkhorshid - MD
simulations, QM calculations
 Shigehiko Hayashi - MD
simulations, QM/MM calculations
Shigehiko Hayashi - MD
simulations, QM/MM calculations
 Jerome Baudry - MD
simulations, Free energy perturbation calculations
Jerome Baudry - MD
simulations, Free energy perturbation calculations
 Ferenc Molnar - QM
calculations
Ferenc Molnar - QM
calculations
 Michal Ben-Nun - Quantum
Dynamics Simulations
Michal Ben-Nun - Quantum
Dynamics Simulations
 Hui Lu - MD Simulations
Hui Lu - MD Simulations



