Purinergic signalling in P2X receptor
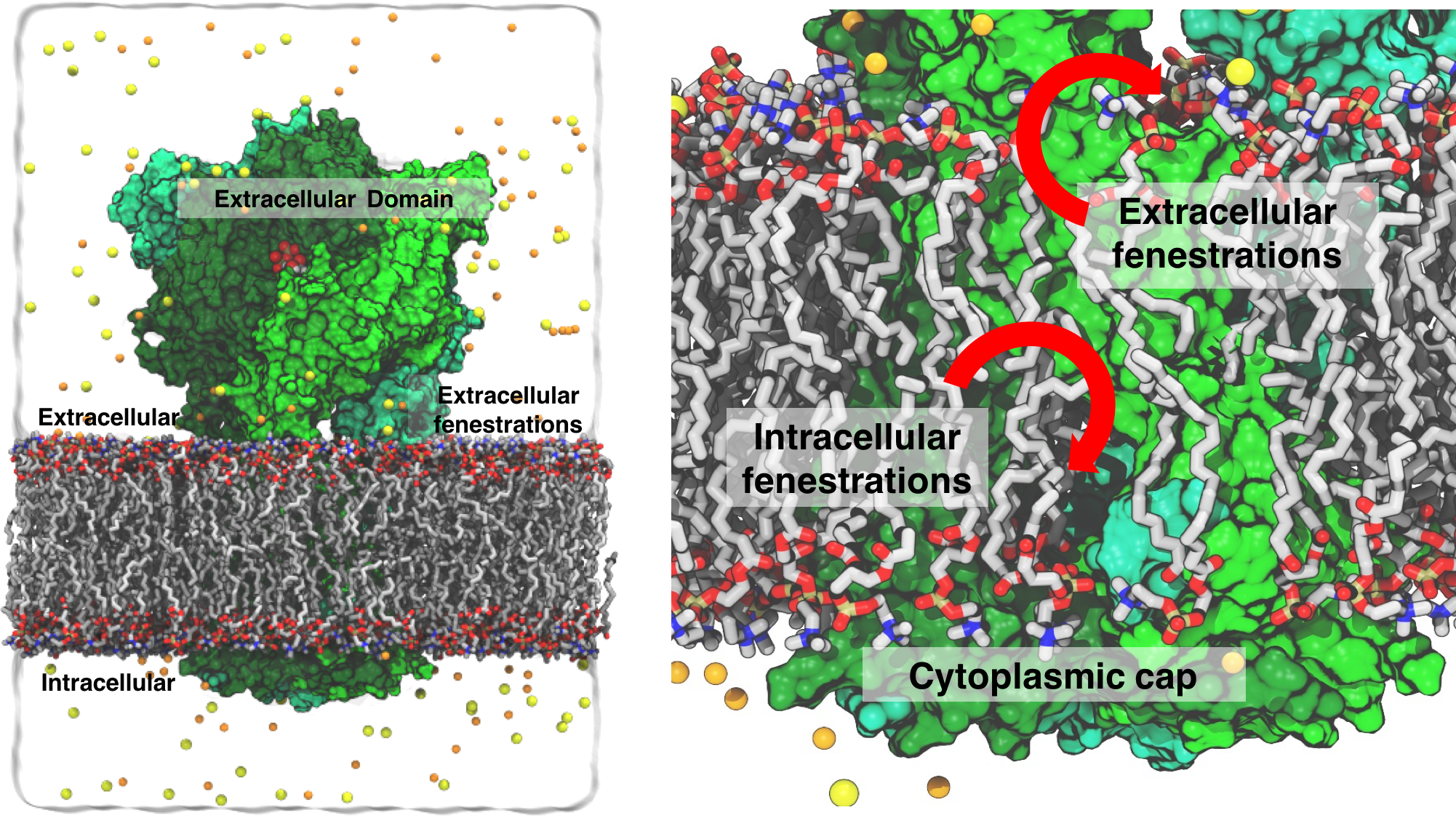
Fig. 1 Simulation box of the P2X3 system. Lipid lining the extra and intracellular fenestrations shown.
ATP as signalling molecule
Adenosine-triphosphate (ATP), a ubiquitously prevalent biomolecule, is not only the principal source of biochemical energy for living cells, but also plays a crucial role in inter-cellular communication, by acting as a signaling molecule. Receptors that bind to ATP and its breakdown product adenosine are widely distributed in the nervous system, among other tissues. These integral membrane receptors are referred to as purinergic receptors signifying the fact that they bind and respond to purines. Ever since their discovery in 1970s, purinergic receptors have been classified in two families, ligand-gated P2X receptors and the G-protein coupled P2Y receptors.
P2X receptors that was the focus of this work are ubiquitously distributed in a wide variety of cells in humans and are responsible for key processes such as muscle contraction, neurotransmission, inflammatory response, pain, and taste signal transduction. P2X receptors are trimeric, non-selective, cation channel that are activated upon ATP binding , thereby playing crucial role in extracellular ATP signaling. Apart from being regulated by ATP, P2X receptors are sensitive to non-competitive inhibitors and are allosterically regulated by metal ions like calcium, magnesium, zinc and copper. Seven mammalian P2X receptors (P2X1-7) have been identified so far, sharing a common topology comprising of homo or heteromeric subunits, each subunit comprising a transmembrane (TM) domain composed of two TM helices (TM1 & TM2) and a large cystine-rich extracellular domain. The TM domain forms a cation pore whose opening is linked to the extracelluar domain where ATP (the agonist), regulatory metal ions, and various antagonists bind. The particular subunit packing of the TM domain, as revealed in the recent crystal structure of P2X3 solved by our collaborator here (Eric Gouaux, HHMI, Vollum Institute), introduces peculiar lateral fenestrations at a level approximately matching the midpoint of the membrane, which are thus expected to be directly lined by the lipid molecules. As such the main question that we addressed was , how would an ion permeate through such a lipid rich environment? Ion access and permeation
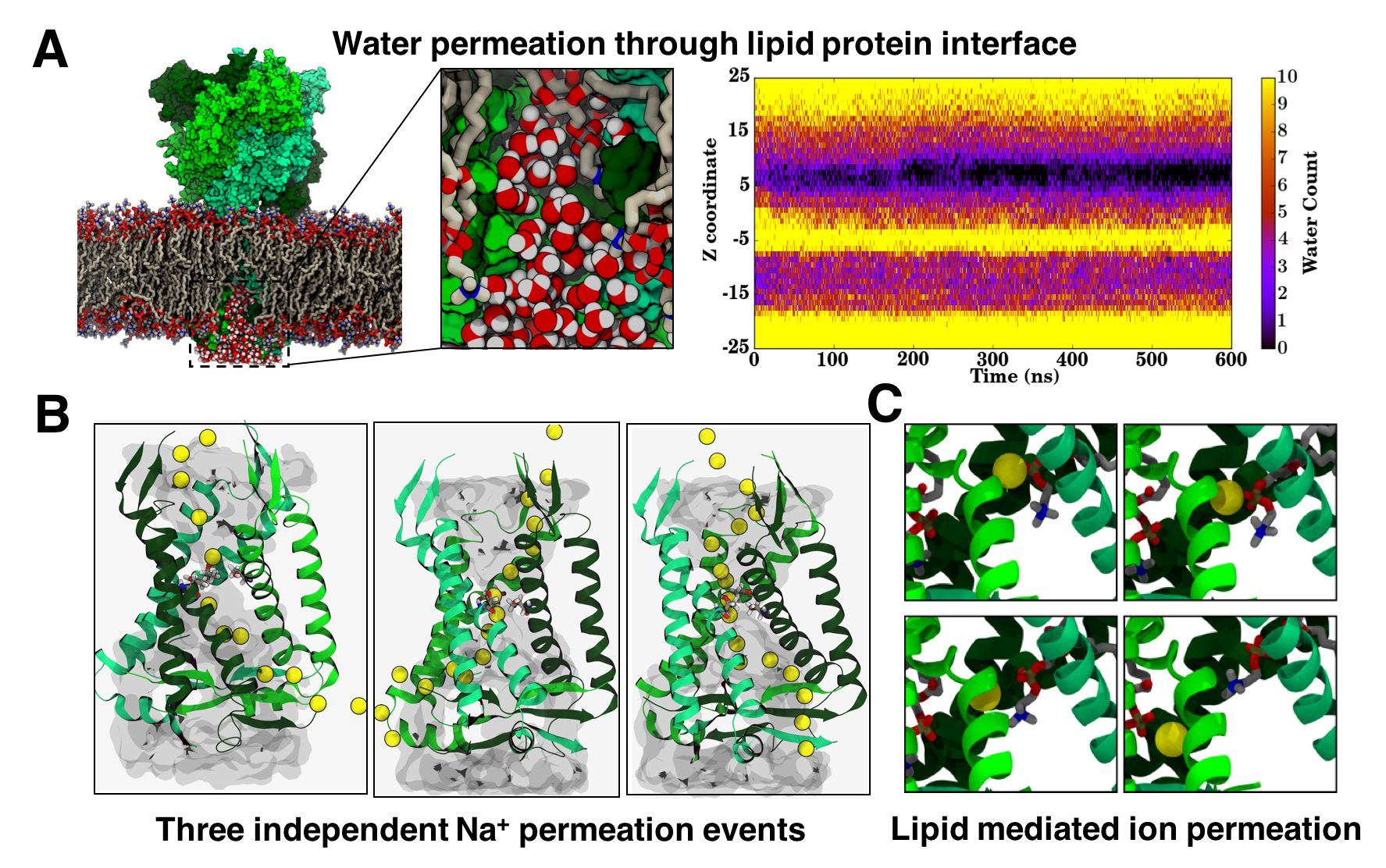
Fig. 2 A) Snapshot showing water permeation through lipid protein interface and water density in z direction for 600 ns.
B) Na+ permeation through all three fenestrations. C) Na+ interaction with POPC as it traverses the channel.
The main objective of this work is to elucidate the Na+ ion translocation pathway, particularly the ion egress mechanism. Although the Na+, enters through the lateral fenestrations of the extracellular vestibule, it is unclear as to how will the ion egress through the protein. Since, the ion cannot egress along the main 3-fold axis owing to the closed cytoplasmic cap being too small to allow for the passage of hydrated sodium ions, the only other pathway possible are the cytoplasmic fenestrations formed at the interface of the two monomers. While these fenestrations are wide enough for the ion to pass, they are directly lined by the lipid molecules presenting unique case of an ion permeation through a lipid rich environment. Hence, in order to probe the permeation of ion through these fenestrations and to study the governing nature of lipid protein interaction on ion permeation, we performed an extensive set of MD simulations of the P2X3 receptor embedded in a POPC bilayer.
Our equilibrium MD simulations show a clear hydration pathway for the solvated ions in the protein core (Fig2: Panel A). These water molecules pass through the open pore but do not exit from the bottom surface of cytoplasmic cap, instead it exits through the lateral fenestrations, through the space between the lipid protein interface (Fig2: Panel A) . Polar head groups of the lipid molecules probably assist in the permeation event. This suggested potential pathway for the ion permeation, however owing to the limited timescales of the MD simulation we could not sample a complete Na+ ion permeation event. In order to accelerate the process of ion translocation, we performed a second set of simulation, in which an external potential of 1 V was applied in the direction perpendicular to the membrane. Application of the external potential of 1 V, resulted in not just the permeation of Na+ ions along the protein lumen, but also the exit through the lateral fenestrations (Fig2: Panel B). Moreover, the permeation was observed through all three fenestrations and along the same pathway taken by the water molecules as seen in the equilibrium simulations. Interestingly the permeation pathway is lined by the lipid head groups, which interacts with the ions as they translocate through the protein lumen (Fig2: Panel C). This work hence shows a unique case where the ion permeation pathway is formed by both the lipid and protein molecule.
Publications
X-ray structures define human P2X3 receptor gating cycle and antagonist action. Steven. E Mansoor, Wei LÜ, Wout Oosterheert, Mrinal Shekhar, Emad Tajkhorshid, Eric Gouaux. Nature , 2016. DOI: 10.1038/nature19367
Investigators
- Mrinal Shekhar
- Emad Tajkhorshid
- Eric Gouaux
Page created and maintained by Mrinal Shekhar.



