Phospholipase
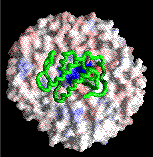 Desolvation of the lipid surface in PLA2--membrane complex.
Click on the picture for a 93k version.
Desolvation of the lipid surface in PLA2--membrane complex.
Click on the picture for a 93k version.
Phospholipase A_2 is one of the most intensively studied membrane proteins which hydrolyzes phopholipids at the sn-2 position to form fatty acid and lysophospholipid products [1]. These are small proteins and the 3-D structures are known to high resolution for several species [2]. Phospholipase A_2 proteins are of high pharmaceutical concern since they are responsible for the release of arachidonic acid from membranes, and since the subsequent conversion of this fatty acid to leukotrienes and prostaglandins is part of the inflammatory response.
The enzyme also shows very interesting interactions with the membrane on which it binds [3]. It is activated in some way when it interacts with aggregated forms of the substrate, such as in micelles or in bilayers. Electrostatic and hydrophobic interactions are suspected to be involved in the binding of the enzyme to the membrane. Very little is known of the enzyme-membrane complex structure and why the enzyme reacts much more efficiently once it binds its substrates in an aggregated form.
We are particularly interested in studying the interaction of the phospholipase A_2 with the cell membrane through molecular modeling and molecular dynamics. THe questions of interest are: the mechanism of the surface activation; the interaction of PLA2 with negatively charged lipids; and the conformational and energetic properties of the lipids in the protein-membrane interface. Our simulations have suggested that a desolvation of 2-3 lipid molecules occurs when PLA is tightly associated with the membrane, and could facilitate the binding of the substrate, increasing the reaction rate. We have also calculated the potential surface of the protein on membrane surface.



