Historical Series: Membrane Proteins
When Membrane Proteins Fuse
with Computational Biology:
A History
By Lisa Pollack
September 2014See other related articles by L. Pollack
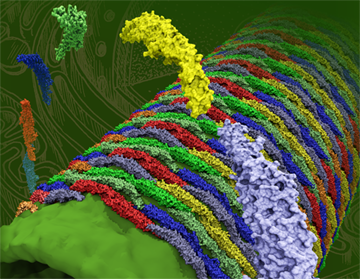
BAR domains, proteins that sculpt membranes.
In the middle of difficulty lies opportunity.
–– Albert Einstein
Klaus Schulten unlocks the mysteries of biology by working in the field of computation, which is a virtual newcomer compared to the well-established and centuries-old realms of theory and experiment. “As a result, everything new is looked at with great skepticism,” says Schulten, “EVEN if it's absolutely necessary like in biology and if it's actually very successful, like in biology.” In other words, his trajectory in the computational sciences has not always been smooth, and he has often met with scorn. He explains that theorists claim computational people are not as intelligent as the theory people, and experimentalists declare that what Schulten does it just virtual reality. In other words, computation is a new branch of science that is having a hard time getting its foot in the door. Schulten explains this is because science is extremely conservative; it has to be, to make sure everything is correctly vetted. So acceptance of this new branch is necessarily slow going.
But why does Schulten persevere as a second-class citizen almost? Very simply, what he does now for a living is what he always dreamed about for his future. He was a kid who loved nature, who loved living systems, but at the same time was mathematically inclined. He always wanted to study living systems, but do it theoretically–using math. And then, early in his career as a professional scientist, he realized that, in order to explain biology and especially its captivating living cells, he needed to use the computer. “Biology is about detail,” Schulten notes. “And to account for detail is really only possible with the computer.” In fact, the intricacies and importance of a biological system like the cell membrane make it a prime candidate for computational studies. “The membrane itself is really wonderful as an object of simulation because the membrane is disordered,” says Schulten. The membrane structure is constantly changing over time, and such rapid fluctuations in structure are very difficult for experiments to capture and see. But not for the computer–it can elucidate the details of the structure as it changes over time. “So this is really one of the best applications of the computer in simulation–to deal with membranes, into which you embed proteins.”
The membrane is where a whole host of fascinating functionality is taking place: inport, export, signaling. Even some chemical reactions occur on the membrane. In higher life forms, for example in human cells, not only is there a membrane around the outside of the cell, but the interior is also compartmentalized by membranes. “It's like a skyscraper with thousands of rooms,” says Schulten of the human cell. “And these walls that separate the compartments are highly functional.” Many of the rooms need to be different from the others, they need to have doors, and some of the rooms even need to be flexible compartments that move through the cell. And don't even get Schulten talking about nerve cells. While membranes are made of molecules called lipids, one might easily expect that dozens of kinds of lipids comprise these complicated cells of the nervous system. But no, in the brain it is more like 300,000 different lipids! Schulten likens these nerve cells to an elaborate billfold with a monumental array of separating sheets all distinct from each other. This kind of cell with its innumerable lipids is only one of the many reasons that draws Schulten toward studies of membranes and the proteins that live in them.
Another reason that these systems tempt Schulten is that he sees them as an opportunity to make discoveries with the computer. A living cell is highly complex. Unlike a physics system such as a crystal that has a unit cell repeating ad infinitum, there is no symmetry to a biological cell. It is more like a country that has societies–neighborhoods, villages, cities, and states. “But the only way I can do it,” says Schulten of studying societies in living cells, “because of this complexity and heterogeneity, is with a computer. So I have this wonderful reward of making discoveries, but the computer is absolutely necessary there.” Membrane proteins are especially fertile ground for Schulten and his love of discovery. Many experimental techniques for studying a protein in a membrane require the protein to be outside of the membrane, that is, outside its natural environment, for example in a crystal. But in computer simulations the protein is embedded in the membrane, to more naturally mimic the original environment of the protein. This gives the computer an edge. Moreover, structure determination by X-ray crystallography often can't capture the protein in its functional state, and so the computer is one tool employed for uncovering a membrane protein active state structure. And, lastly, since computer simulations chart behavior over time, Schulten can often see the protein in action, that is, observe the protein perform its function, whether that be watching how a pore selectively allows water through to the exclusion of protons, or witnessing how membrane proteins actually sculpt its membrane into a desired shape or compartment.

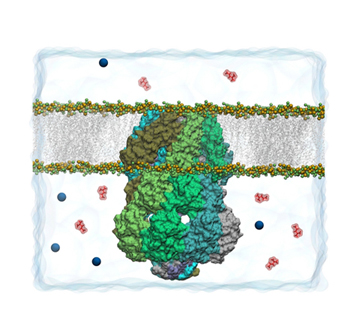
A spinach aquaporin, a membrane protein that conducts water, one of the many membrane proteins Klaus Schulten has tackled in his career.
Klaus Schulten began his research career in 1970, but literally had to wait almost thirty years to study membrane proteins, although they were always on his mind in his early years. He needed the actual structure of the protein to make any significant discoveries, and he needed to hone the computer to a point where he felt confident that this tool was sophisticated enough to handle protein + membrane. No one was able to crystallize membrane proteins to yield a structure until 1984, and then only slowly were more structures forthcoming. Now, in the second decade of the twenty-first century, Schulten has an array of membrane protein structures to work with, and this has become somewhat of a hot field. For good reason. Estimates are that about a quarter of human proteins encoded by the genome are membrane proteins; and over fifty percent of pharmaceutical targets are membrane proteins.
But Schulten's path in this field has not always been easy. While Schulten has elucidated many membrane proteins, he had to overcome obstacle all along the way to get results–not just the usual ones facing a computational scientist. He realized very early on that computers were going to become essential in biology, but could get no one to work in computational biology with him when he was a young professor. The result was a stunning and outrageous plan he concocted to see for himself if the computer was suitable for characterizing membranes. He also had to figure out the best way to employ parallel computing in his research when parallel computing was in its infancy, which required another intrepid plan of action. And his pioneering work on one of the largest systems at the time was questioned at every turn.
Yet, such adversities were coupled with perseverance, and Schulten only got better and better at using the computer for biological systems. And his odyssey with membrane proteins mirrors his research group's expanding finesse with large computer simulation. The goal of such large calculations for Schulten is to describe biological organization, to explain protein cellular societies, for the purpose of making discoveries. When Schulten was a young scientist, he never dreamed of the progress he and others would make on membrane proteins. “If somebody would have told me you will never be able to describe the membrane and the protein in one, I would have said, 'Too bad, maybe I should switch, but I find it so interesting, so probably I won't,'” he reflects. Luckily for Schulten, his persistence paid off and his history with membrane proteins is rich and has given him many opportunities for discovery. Schulten says professionally he is constantly driven to “turn over a stone and look what is under it, and open a new door and peek into a new room.” The fusion of membrane processes with the computer surely has given Schulten many new rooms to view, as we shall see below.
Membrane Proteins: Always on the Horizon for Schulten
By the mid 1970s Klaus Schulten had an array of interests in biological systems that he knew, unfortunately, he could not study with any certainty. And he wasn't even sure if it would be possible, later on down the line, to examine a protein whose natural home was a membrane environment. “So one couldn't really see how far one would make progress,” Schulten reiterates. Nevertheless, he was fascinated with processes dependent on proteins in membranes: photosynthesis, vision, and bacteriorhodopsin-based bioenergetics. For the kind of studies Schulten wanted to do, he needed the structure of membrane proteins, and while many people were studying these systems at the time, for a physical biologist like Schulten, there was too much uncertainty without the structure on hand. Textbooks at the time even purported that it was impossible to make crystals out of membrane proteins, which would be a necessary first step in using X-ray crystallography to unveil a high-resolution structure.
His fascination with vision and photosynthesis can even be traced to his boyhood. When he was still a young boy he read a series of science books for children, with titles like [You and Mathematics] or [You and Chemistry]. But the children's book that most intensely captured his imagination was by the acclaimed Austrian ethologist Karl von Frisch, entitled Du und das Leben [You and Life]. Although Schulten most likely read this book 50+ years ago, he still vividly remembers the chapter about how life forms make ATP through either photosynthesis or food consumption. “I felt that was my calling,” asserts Schulten. “I wanted to understand how life forms use energy to make ATP and then run many processes with it.” In fact, Schulten's take home message from the book was that to understand life, one needed to explore the underlying molecules that form living cells, as von Frisch stressed in his book. While the molecular study of life forms was in its infancy in the early 1960s when Schulten first read this book, it piqued Schulten's interest enough to set a goal for his teenage self: uncover intellectually, as opposed to experimentally, the atoms and electrons that make up the molecules of life. Remarkably, that is exactly what he does today.
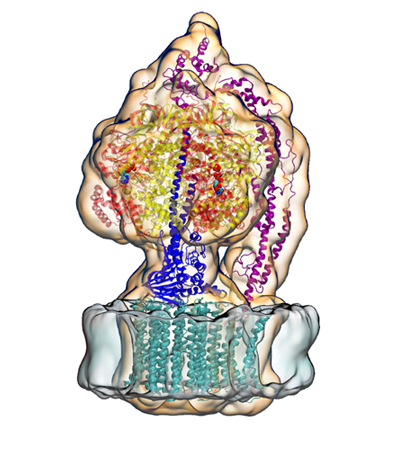
ATP Synthase, a membrane protein Klaus Schulten would eventually study to help fulfill his childhood dream of understanding ATP, the universal currency of life.
Schulten's high school days were obviously filled with outside reading, but he did this because he was so bored with the topics his instructors were trying to teach. In fact, he regularly challenged his teachers with his outside reading and for this he was nicknamed “Professor” by the other students; they never called him Klaus. In one such tense interaction, the teacher called on Schulten to give a presentation from textbook reading about the lenses in the eye, as no one else would raise their hand. It was clear the teacher picked Schulten only as a last resort. After Schulten covered the textbook information on lens behavior, he started telling the class how light is received in the eye, and about rhodopsin (a membrane protein, incidentally). The teacher was furious because this last bit was not in the textbook. Schulten received a verbal upbraiding, but he was used to this and relays it didn't bother him.
While vision and photosynthesis may have captivated teenage Klaus Schulten, his understanding of vision grew much deeper when he went to Harvard University for graduate school in physics 1969. There in Cambridge he sat in on the group meetings of pioneer George Wald, who won the 1967 Nobel Prize for his studies on visual processes. From Wald, Schulten learned about the cutting edge work on rhodopsin, a membrane protein.
In the human eye, the photopigments that absorb light are in the rods and cones. These photopigments are comprised of proteins that bind retinal, the visual chromophore newly described by Wald. In the rods, for example, retinal is bound to the protein rhodopsin, while in the cones it is bound to the protein photopsin. The rods and cones function in the same way, except that the wavelengths of absorption are different, but both rely on the core molecule of retinal. One side of a long rod or cone is composed of a series of flat disks stacked on top of each other, with the retinal + protein sitting in the membrane of these disks. In a rod, when light strikes the photopigment, retinal changes shape, and then a series of steps ensue to convert the shape change to a nerve impulse. While rods absorb in one band, cones are either red-, green-, or blue absorbing; small changes in the opsin proteins determine the different absorption peaks in the cones–once again, retinal remains the same in the three different visual receptors of the cones.
Retinal is amazing because it is the core molecule of vision in sighted species, even though eyes evolved independently in the different phyla insect, mollusk and chordata. George Wald uncovered that retinal + opsin was the photopigment responsible for vision, and realized that retinal is a form of vitamin A. He was in fact a central figure in studying the molecular aspect of vision–an aspect that intrigued young Schulten, who wanted to study for himself the molecules of life. And Schulten to this day remembers all the fascinating details Wald shared about retinal; to name one, Schulten recollects that Wald explained how bullfrogs have two kinds of retinal, with one working more when the animal gazes at the blue sky, and another for looking at water, a reddish scene. In this instance the length of the retinal is minutely different, so that it absorbs either in the blue (sky) or red (water) depending on the needs of the animal. A bullfrog might need to gaze at the sky for circling predator birds, or might gaze down at the water for an insect meal to land. “To me it was fascinating,” Schulten says about George Wald's anecdotes, “because I studied retinal, and to realize there are two retinals and that one is absorbing a little bit more in the red. And nature uses it actually.”
While vision and photosynthesis had always been on Klaus Schulten's radar when a graduate student, he got a chance in a 1978 publication to actually work on photosynthesis. While this cemented for him a lifelong love of photosynthesis, he would wait ten years for the next opportunity to study this topic, for the key proteins that participate in the process are membrane proteins. In 1978, with no structure of any membrane protein yet determined, much less the proteins involved in photosynthesis, Schulten could not really delve deeply into this topic to a degree that he felt comfortable. But the observation he made in his 1978 paper was a helpful contribution to a field.
Retinal (in red), a universal molecule in all sighted species, whose many properties Klaus Schulten first learned about from pioneer George Wald.
By 1974 Schulten had finished graduate school at Harvard and he landed his first job in Germany, at the Max Planck Institute for Biophysical Chemistry. He was literally the sole theorist in an experimental group run by Albert Weller, a director there in Göttingen. Weller immediately gave his new theorist a conundrum to solve: figure out why a product in a standard electron transfer reaction appears so much faster than expected. The product was a an excited molecule with two electrons whose spins were parallel; this product was coined a “fast triplet,” for in the language of quantum mechanics two electrons with parallel spins are known as triplets. Schulten set to work and unraveled why fast triplets were occurring in Weller's pet reaction.
One of the important things he uncovered from that exercise was that a magnetic field could affect a chemical reaction, which in the mid 1970s, was a topic studied by some but many doubted its feasibility. Basically Schulten realized that to prove his claim, experimentalists could run a reaction both with and without a magnetic field present and corroborate his theoretical finding. For suggesting this, that a magnetic field can alter a chemical reaction, Schulten was the laughing stock of the laboratory, until some experimental data from group member and good friend Hubert Staerk confirmed that Schulten was not crazy.
One of the significant things that Schulten postulated from working on the fast triplets mystery was that he realized that a certain kind of molecule (one that is capable of forming radical pairs), could act as a compass in a bird for detecting a magnetic field. This story, of Schulten postulating that a specific molecule could account for avian magnetoreception, is told in greater detail in this article. Suffice it to say, Weller's dilemma provided Schulten with an excellent problem to solve for many rich returns on investment.
In fact, it was Schulten's observation about a magnetic field effect on a chemical reaction that brought him briefly into the field of photosynthesis while he was in Göttingen. Weller's group was focused on electron transfer reactions. Weller's pet system was a chemical system (pyrene and dimethylaniline), but Schulten says everyone in the group, including himself, was aware that electron transfer reactions also happened in biological systems, namely in photosynthesis. But the exact details of the electron transfer in the proteins responsible for photosynthesis were not known exactly–to know that would involve discerning the high-resolution structure of the photosynthetic membrane proteins. “So people knew, however,” explains Schulten, “that there is a special pair of chlorophylls that absorbs the light and that then transfer an electron step-wise through these proteins that sit in a membrane and thereby charge the membrane.” While other scientists, including some in Weller's group, had pieced together models for the key groups of chlorophylls, as well as some detail about the pheophytins and the quinones, one couldn't be certain how everything was exactly working without a structure. At this time of uncertainty Schulten says that every additional observation about photosynthesis and its accompanying electron transfer reactions would be helpful.
Enter Schulten and his discovery that magnetic fields can affect a chemical reaction. Schulten realized that magnetic fields could affect a biological reaction exactly like the electron transfer ones seen in photosynthetic organisms like plants and bacteria. Schulten's first ever publication about photosynthesis was basically a suggestion to the scientific community that an experiment done both with and without a magnetic field would yield an observable that could be measured. The observable: triplets. So an experimentalist could notice a difference in outcome based on the presence of absence of a magnetic field. Schulten says that everyone in Weller's group was aware of biological systems that experienced electron transfer, so the leap to suggest that experiments could measure triplets in photosynthetic electron transfer was an obvious one. He published this suggestion in 1978, working with a student he was mentoring, Hans-Joachim Werner, who later became a world leader in quantum chemistry.
The Literal Red Herring
“It was under their noses all the time,” Schulten says about a bacterial system that fascinated him in the 1970s, when its important membrane protein was discovered. While Schulten was intrigued by the visual receptor rhodopsin at least as early as high school, and then grew more deeply appreciative of its role from George Wald, all the rhodopsins known at the time belonged to the animal kingdom. Could it really be that a bacterium had a rhodopsin, as a new discovery in 1971 suggested? Bacteria don't have sensory proteins. What was a rhodopsin-like protein doing in its membrane?
Schulten relates that the bacterium in question, called, at the time, Halobacterium halobium, indeed existed in various environments but scientists didn't realize it. “One environment is actually the proverbial red herring,” claims Schulten. “The red herring is the bacteria-infested fish.” In earlier times, people regularly salted fish to preserve it, because bacterial contaminants can't reproduce in salty environments. What kind of bacterium is this that lives in the preserved fish then, literally in salted herring? Turns out, it isn't a bacterium at all. It's a type of extremophile called a halophile (Greek for salt-loving), which belongs to the domain of life called archaea. But when Schulten first heard that a rhodopsin protein was found in halobacteria, in the early 1970s, everyone assumed these salt-loving organisms were bacteria. In fact, to make matters more confusing, the rhodopsin protein was named “bacteriorhodopsin”, a name that remains even to this day. (While it was discovered in 1971 that there was a rhodopsin-like molecule in H. halobium, which was later filed under archaea, in 2000 a group of scientists did indeed find a bacterium that contained a rhodopsin protein, so it is not just archaea that have these proteins. All domains of life can now claim to have their own retinal + opsin complex.)
The reason the halobacteria were considered to belong to bacteria was that the revolution that shook up the tree of life did not occur until 1977. This revolution supplanted the two domains on the taxonomic tree with a three-branch revision. Carl Woese, then a scientist in the department of microbiology at the University of Illinois at Urbana-Champaign, realized that the unique organisms he was studying, methanogens, did not belong to the bacteria domain. For years Woese had been trying to classify bacteria by the sequence of their ribosomal RNA, instead of with physical features. When a colleague down the hall was discussing the microbes he was studying, Woese decided to sequence these organisms that came right from the cow intestines and cow manure at the scientists' land-grand university there in Champaign-Urbana. Woese immediately recognized that their sequences were not like any other bacteria he had studied, and he had studied quite a few. Although he then proposed a new domain of life, the archaea, in 1977, his finding was met with scorn. That story, of Woese and the eventual acceptance of his idea, is told in many other sources. Suffice it to say, when Klaus Schulten heard about bacteriorhodopsin in the mid 1970s, everyone was under the impression that it inhabited a bacterium. Woese's proposition did not really catch on until the 1990s, and even today some scientists don't embrace the three domains of life: bacteria, eukaryotes, and archaea.
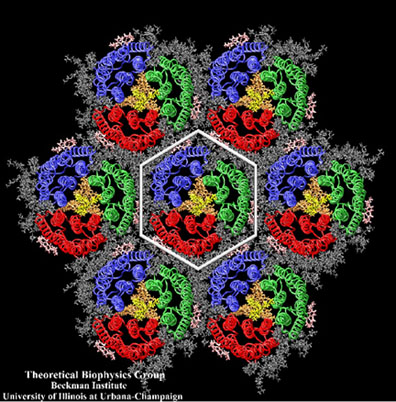
The crystalline-like purple membrane, a system Klaus Schulten would go on to study in the late 1990s.
Scientists studying the salt loving Halobacterium halobium in the late 1960s, namely with efforts led by Walter Stoeckenius, realized from experiments in the laboratory that the cell membrane of this organism could be broken into fragments, and that studying the fragments would be easier than studying the intact entire membrane. They found that the fragments would naturally separate into functional patches based on protein content. The most intriguing fragment was purple, and grew to have the designation “purple membrane.” It was compelling to scientists for many reasons. First, the proteins of the purple membrane are arranged in a regular array. Solid state physicists would call objects that have repeating identical units crystalline. Indeed, Stoeckenius found the purple membrane to be a two-dimensional hexagonal crystal. Second, only one type of protein exists in the purple membrane. A system this basic is always on the radar for scientists, as simple systems allow them to characterize processes otherwise unattainable in more complex life forms that undergo those same processes. Hence, a bountiful amount of data has accumulated over the years on the purple membrane, a bonanza model-system for scientists (and Klaus Schulten even initiated studies of the purple membrane in the 1990s). And lastly, the lone protein in the purple membrane is bacteriorhodopsin. Again, what was a sensory protein doing in a bacterium?
Walter Stoeckenius wrote a 1976 Scientific American article where he details the discovery of the purple membrane and bacteriorhodopsin. Although he is not certain the exact reason why his group thought to look and see if this halophile had a rhodopsin-like protein, he points to two possible reasons. First, another group member had previously studied the photoreceptor membranes in the eye, and the whole Stoeckenius group was aware of this previous research. And, second, Stoeckenius had noticed something unique over the course of his many studies; the cells in solution swam in a different direction when red light was reduced. Using the established procedure for studying rhodopsin, group member Dieter Oesterhelt indeed found a retinal, and they named the photoreceptor bacteriorhodopsin. The men published this result in 1971. Stoeckenius then describes in Scientific American the lengths he and Oesterhelt had to go through to learn that the bacteriorhodopsin functioned as a proton pump, which they published in 1973. So the mystery was solved. Bacteriorhodopsin was creating an electrochemical gradient between the cell interior and the exterior. Stoeckenius then went of to prove that this proton gradient was used to synthesize ATP.
It seems obvious why discovery of bacteriorhodopsin in the purple membrane intrigued Klaus Schulten in the 1970s. Here was a primitive organism, a bacterium–at least so envisioned at the time–that had a sensory protein. The fact that it was more primitive than the human eye meant it was a simple system for scientific study; as stated above, scientists love simple, model systems. But perhaps most intriguing for Schulten would be that bacteriorhodopsin was central in ATP production. After all, Schulten felt like it was his calling in life to understand ATP. Schulten already knew that in photosynthesis, light energy is converted first to electron transfer and then proton transfer. The proton transfer in photosynthesis was used to make ATP, much as the proton transfer in bacteriorhodopsin drove the formation of ATP in the halophile. Hence, the three membrane systems, rhodopsin, bacteriorhodopsin, and the photosynthetic proteins, all were on Klaus Schulten's mind in the 1970s. He would have loved to study all of them further, but no high-resolution structure existed for any of them. It didn't look like any structure would be determined any time soon either. For example, in a 1977 review article on the purple membrane, by Richard Henderson, he laments all the unanswered questions that just need a high-resolution structure. And then goes on to write
Clearly, the answers to these questions will come from a knowledge of the structure of the retinal-protein complex and the sequence of conformational changes that follow the absorption of light. These changes in conformation will presumably include changes in the positions of the atoms in retinal. However, the detail necessary to see individual atoms is unlikely to come from diffraction methods.
The diffraction method he is referring to is X-ray diffraction or X-ray crystallography, at the time the only known way to obtain high-resolution structures. This quote gives some of the idea of the prevailing ideas at the time, in the 1970s, about membrane protein study. But all that was about to change in the early 1980s, and Klaus Schulten would have a front row seat to the excitement. And it would prove to have lasting consequences for his future.
Resolute in Bavaria
Talk about being in the right place at the right time. For a young scientist who was fascinated by photosynthesis, but realized he could do nothing without a structure, Munich in the 1980s could not have been a better choice. There in Bavaria Klaus Schulten got a permanent position at the Technical University of Munich in 1980; and while a professor he got to know Hartmut Michel, who worked in the next town over at the Max Planck Institute for Biochemistry. Michel was going against all known wisdom and attempting the impossible: crystallizing a membrane protein. And, probably to the delight of Schulten, Michel was attempting his improbable feat on a membrane protein central to photosynthesis, the reaction center.
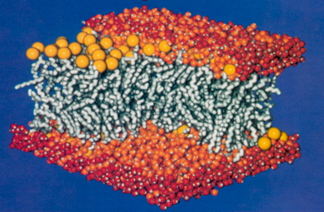
Since the 1950s scientists have been using X-ray diffraction to determine high-resolution structures of proteins; requisite to this technique was the formation of the protein into a crystal, such that perhaps a billion proteins were grown together to form an ordered array, a crystal. But making protein crystals had only been done for proteins that normally live in the cell's cytosolic fluid, i.e., an aqueous environment. When Hartmut Michel began his quest to crystallize a membrane protein in 1977, textbooks at the time purported it was impossible to make a crystal out of a membrane protein. This is because of the properties of the surface of the proteins. Basically, a membrane is comprised of a lipid bilayer, as shown. The lipids look like balloons with strings attached. The heads of the balloons are water loving, and are exposed to the watery inside and outside environments of the cell. But inside of the membrane, where the long tails reside, is a hydrophobic environment. So the majority of the surface of a membrane protein lives in the water-hating environment of the tails, and as a result the membrane protein surface is usually nonpolar. But a cytosolic protein usually has a surface that is water loving, and it may be either charged or polar. As a result of their electrically polar surface patches, cytosolic proteins actually interact very strongly with each other so they are much more likely to form crystals than do membrane proteins.

A typical cross section of a lipid bilayer.
But something had to convince Hartmut Michel that he could indeed make crystals out of a membrane protein. Enter bacteriorhodopsin. As noted above, Dieter Oesterhelt was one of the scientists who discovered bacteriorhodopsin and that it acts as a proton pump. In 1974 Hartmut Michel joined Oesterhelt's group and started his own studies on bacteriorhodopsin. In June of 1977, while working on a project, Michel noticed that delipidated bacteriorhodopsin in the freezer formed glass-like aggregates. This gave him the idea that indeed it would be possible to make crystals from a membrane protein. Dieter Oesterhelt was completely behind this risky endeavor Michel wanted to implement. Accordingly, Michel then tried various types of membrane proteins in his quest. In point of fact, by 1981 he had made excellent quality three-dimensional crystals out of a reaction center protein. (Making crystals was one thing; making crystals suitable for X-ray diffraction was a whole other ball game.) He joined forces with an expert in X-ray diffraction there at the Max Planck Institute for Biochemistry, Johann Deisenhofer, and by 1984 they published their first structure. They were awarded a Nobel Prize for their work in 1988. A more detailed account of Hartmut Michel's efforts to crystallize a membrane protein is given in his autobiography on the Nobel Prize website.
So what did Hartmut Michel actually do to form crystals? The simple answer is: use of detergents. These are lipids, but with short tails instead of long ones. He made crystals out of a membrane protein with the help of soap, as soap contains detergents. His choice of protein, the photosynthetic reaction center, was also a very scientifically relevant one. He crystallized the protein that came from a purple bacterium, as plants are not the only living organisms to practice photosynthesis. The reaction center is the core protein in the process of photosynthesis; there light energy is captured or auxiliary proteins funnel their excitation to it. Then that excitation energy is converted into electron transfer. Schulten, who witnessed much of the work of Michel and Deisenhofer, says that the pair was lucky because the reaction centers of the crystal functioned exactly like they did in the membrane. Namely, when shining a light on the reaction center crystals, they behaved optically just as they did in the membrane; they were more or less completely functional. Not only did the perseverance of Michel and Deisenhofer ignite the field of membrane protein analysis with their finding, it was also a clear breakthrough for many studying photosynthesis. In fact, the impact on Klaus Schulten was monumental, indeed, and would led to a brash scheme even he admits he might not have followed on second thought.
Inelegant Computing
“For a strange reason I did my second goal first, because I had to put the protein somewhere,” Klaus Schulten says about the effect the reaction center had on him. “And somehow it seemed more natural to me, that I first would do the membrane piece.” By 1987 Schulten realized that structure of the first membrane protein, whose solution happened right under his nose, would offer him new and unexplored vistas as a theoretical biophysicist. He was delighted, but confounded at the same time. In the late 1980s, the biggest protein simulations were about 3,000 atoms. The reaction center was roughly 10,000, a whole order of magnitude larger than what current state-of-the-art could (barely) handle. Goal one: Go big–not the small stuff of just a few thousand atoms. But the reaction center lived in a membrane, and to properly simulate that companionate system, Schulten needed to embed protein into membrane. In fact, embed it in a large piece of membrane, as just a few lipids would not give a realistic picture of what the membrane protein saw. Goal two: simulate a big enough patch of membrane.
What Schulten realized, at that point, was that to accomplish his new goals, he would need a supercomputer. But it would have to be dedicated to his task for years, to the exclusion of anyone else in the queue. At this point in time, the existence of supercomputers made available for ordinary scientist researchers (as opposed to defense workers or industry) was merely in its infancy. It was only in 1985 that the National Science Foundation chartered the program to build national supercomputer centers for the express use of academic researchers. So, while Schulten knew about supercomputers, it wasn't a staple in the arsenal of the average scientist in 1987.
While access to supercomputers may have been rare in 1987, and something only in the back of Schulten's mind, he was, however, keenly aware of computing as a tool. A tool he couldn't fully utilize. During his time in Munich in the decade of the 1980s Schulten became more and more aware that, to fulfill his dream of studying biology via physics and mathematics (instead of in an experimental laboratory), he would need to account for the detail inherent in biology; and to do that, he would have to steer his research toward computational biology. But he didn't think his own finesse with computing was first rate–in other words, he didn't live and breathe computers, the way ardent enthusiasts sometimes do.
Biology is about the detail.
“I begged other people to do it who knew more about computers, but they were too much in love with theoretical physics and didn't want to do something as inelegant as computing,” says Schulten of his attempts to employ computation for biology. The main people he asked to undertake computational biology were his students. They were all from the physics department, as back then in Germany it was almost unheard of for a student from engineering or chemistry to cross fields and work with Schulten in physics. To be clear, Schulten and his physics students did use computing in their work, but it was nowhere at the level as is used today, and what they did certainly wouldn't be classified as computational biology. Instead, his physics students wanted to be great theoretical physicists and thought computing was primitive. “All my students…wanted to become the next Einstein,” Schulten jokes about his time in Munich. “They were not wearing socks with sandals because Einstein wasn't wearing socks and so they weren't wearing socks.” Nevertheless, Schulten continually told everyone in his group: Do your future with computing. But his students resisted, claiming it wasn't elegant enough for their own tastes.
To this day, Schulten can't figure out why all his theoretical physics students, and for that matter, the other professors in his department, looked so unfavorably upon computing. “I don't understand it because to do really great programming, you have to be very intelligent, and it's just mathematics. So, whatever great mathematics you can do, you can often express it in a computer algorithm and do it.” But the message was clear–Schulten would have to do the inelegant tasks of computing himself, unable to rely on the younger generations who had a natural affinity for this burgeoning technology. With the solution of the reaction center, Schulten was really desperate to get to work on his two big goals: simulation of large protein and simulation of large membrane patch. And he was trying to figure out how he could possibly accomplish this without a natural prowess he believed he didn't possess and without a supercomputer all to himself. And then one day in Munich in 1987 during Schulten's office hours, in walked Helmut Grubmüller.
Risking it All
Helmut Grubmüller was a physics student at the Technical University of Munich and fascinated with computers. So fascinated, in fact, that in his second or third semester, around 1986, he built his own small parallel computer. Of course, at the time, he didn't call it a “parallel computer,” just a multi-processor machine. He had his own (serial) computer, although this was in the days before PCs were ubiquitous, and he took it apart and played with it to learn what he needed. But to build his own machine, he soldered his own electronic circuits and bought chips from the manufacturers, learning everything he could from the chip data sheets. He even talked to technical representatives from many companies to learn esoteric details he needed.
“And that very quickly blew my budget,” Grubmüller explains about his small multi-processor machine. “My private budget was like a thousand dollars or so.” But then he concocted an admittedly crazy plan. Like Schulten, he wanted to “go big.” He was not satisfied with the small parallel computer he built on his own. So he decided to approach all his professors and propose the following: he would build them a large multi-processor machine and they would pay for the hardware. “That was my naïve idea,” reiterates Grubmüller. “I didn't want to get paid for that, I just wanted to have the hardware paid, such that I could fulfill my dream–building a larger, faster computer.” All the professors he approached rejected his plan outright.
However, Klaus Schulten remembers the day Grubmüller walked into his office in the Physics building at the Technical University of Munich. Grubmüller actually had brought his small home-built computer in a bag, and showed it to Schulten. He explained a bit about what it was capable of and demonstrated some calculations. As Schulten remembers it, “I almost fell off my chair!” Schulten immediately incorporated Grubmüller into his group.

Klaus Schulten, Helmut Grubmüller, and Helmut Heller and one of the boards from their homemade parallel supercomputer, circa 1988.
Around this same time that Grubmüller was trying to build a bigger, faster computer, another student in one of Schulten's courses had given a presentation on fractals. But instead of the usual routine–lots of talk, equations and maybe a transparency–this student brought in a computer he had rewired and reworked to make it capable of making a movie, which was incredibly sophisticated tinkering on a primitive machine that could barely display a static image. During the presentation, the student opened the back of the computer and explained what he did to make possible showing of a movie. Schulten was immediately impressed and asked this student, Helmut Heller, to join his group. Finally, Schulten had students who were interested in computing. The constellation of factors all came together at the right time: two fascinating science goals, two students who loved computers, and promise of a fast computer. Perhaps that might explain, in some small part, why Schulten's next move was the riskiest of his career. He handed over ALL his grant money to the two Helmuts to build him a parallel supercomputer.
It is impossible to fully convey why the gamble Schulten took was high stakes. Although the two students were more than competent–Schulten had seen that first hand–what they were proposing had never been done before. Supercomputers at the time were millions of dollars. Schulten's grant money amounted to 100,000 Deutsche Marks, about 55,000 US dollars. If somehow the machine failed, Schulten would have nothing to report to the grant agencies, and no results to publish. This might jeopardize his whole career. But he explains that the two young students thought in terms of solutions rather than problem. Anyone who knows Klaus Schulten understands that he has a great appreciation for young talent and for initiative. “I believe in people. And when I believe in people I let them go,” Schulten comments, somewhat explaining why he took such a monumental risk. Suffice it to say, the story of building a homemade parallel supercomputer is covered in great detail in this history, but the students did eventually succeed in crafting a working machine after great exertion and creative problem solving.
Would the Computer be Suitable for a Membrane?
Upon completion of the homemade supercomputer, Schulten's next step was a long journey whose final destination was the great unknown. It would be long because Schulten and his students realized they would need to run the computer for about three years. It would be unknown because Schulten had no way to confirm if the computer was suitable for describing membranes. Again, this was another big risk in Schulten's world. What if he put all this time, effort, and money into one single calculation and the results were rubbish?
Schulten essentially knew that, since it looked like more structures of membrane proteins were on the horizon, eventually he had to describe a system of protein embedded in a membrane. But he decided to dedicate his homemade parallel computer to just looking at a large piece of membrane. Up to that time, only small patches of membrane, that is, very few lipids, had been simulated.
The reason Schulten needed a large swatch of membrane is because of the surface-volume issue. Imagine the membrane patch as a box, and if the box is small, the system is dominated by surface. The bigger the system is, the more volume you have relative to the surface. So taking a box big enough was imperative to get the volume-surface ratio just right to compare to experiment. Schulten was aware of others' work on simulating lipids, but really wanted to be sure for himself that the computer could do a good job describing this system. He was afraid the papers he read on membrane simulation had overstated their claims of success. It is hard to convey how many unknowns Schulten faced in this entire project.
A 200-lipid patch of membrane plus water, the system simulated with the homemade parallel supercomputer.
The type of calculation Schulten and his students were determined to run on a large patch of membrane is called molecular dynamics. This type of simulation charts the behavior of the atoms in a system over time–it essentially gives a dynamic view of systems like proteins or membranes. As such, the calculations need to cover the unfolding of events in a time period. Back in 1990 when Schulten ran this membrane system, he was only able to cover the time period of 263 picoseconds. This is almost a joke by today's standards (where hundreds of nanoseconds is standard, and some researchers can even get to a millisecond for small systems). But even with a state of the art computer all to himself, this was the longest time he could possibly simulate. They ended up doing 200 lipids, with 100 in each layer, and altogether there were over 23,000 atoms in the system, including water.
Schulten fully admits he and his students didn't work any miracles. They used conventional molecular dynamics (and its accompanying force fields)–and didn't purport to invent anything new. From nuclear magnetic resonance spectroscopy experiments they knew the orientation of the bonds that make the lipophilic chain of the lipids and they could calculate this from their molecular dynamics simulations. After three long years of running the homemade computer, they finally got results. Much to Schulten's delight, the simulations agreed with experiment!
“That meant that the computer described the membrane well, exceedingly well actually,” emphasizes Schulten. While maybe not a groundbreaking discovery in the world of science, it was a real eye-opener for Schulten. He saw that the computer was really strong in describing a disordered material like a membrane. Schulten now had his doubts assuaged; the computer could really handle something that mattered–the membrane.
Schulten's celebration over the computing success was short lived, however. When he sent his article on the large membrane calculation to the Journal of Chemical Physics for publication, it was rejected within a week. In fact, he was so mad, he refused to send to that journal ever again for years, But he did quickly publish the work in another journal, and this stands as a highly-cited article for him, to this day.
They Won't Lie to Innocent Young People
In 1992 Klaus Schulten was facing a huge dilemma any scientist would be grateful to have. How do I best spend half a million dollars on equipment? It was 1992 and Schulten had just gotten word that he was going to receive funding from the National Institutes of Health (NIH) to start a computational biology center. He had first endured a two-year trial period with a small center, and when that went well, he was given start-up funds in 1992 to last for a five-year period. Schulten thinks a large part of his successful proposal was due to the homemade supercomputer his group had built, which clearly demonstrated the role computing could play in biology.
As mentioned above, in 1985 the National Science Foundation authorized start-up of four places across the United States to become supercomputer centers, to give average researchers access to these powerful machines. One of them happened to be at the University of Illinois, the school where Schulten was working when he got the green light to go forward with his NIH-funded center in 1992. In fact, the presence of the Urbana-Champaign supercomputer center is one of the reasons Schulten took his new job in the United States. He liked the focus on computing in Illinois.
When he described his equipment money dilemma to the supercomputer people there at the university, they had a suggestion. Use your money to help us buy a-state-of-the-art supercomputer called the Connection Machine; you'll have plenty of access to it but we'll also need two of your five NIH center positions to facilitate the deal. Schulten could have never afforded to buy his own Connection Machine, as it cost upwards of five million dollars.
Schulten contemplated this as an option. His group had already used a Connection Machine to do some research on neural networks, among other things. But he needed to spend his money on a machine to run molecular dynamics simulations–completely different from neural network research and calculations. Schulten reflected on everything he knew about the Connection Machine from talking to lots of people. He seriously considered the Connection Machine as a choice, but he was skeptical. “I had great suspicion of these computers,” Schulten recalls, “because I heard them always being described from a very esoteric point of view, and not from a very practical point of view about what you could really compute with them.”

An image of the first homemade supercomputer Klaus Schulten built. His group would go on to build many more parallel clusters.
In retrospect, Schulten summarizes the dilemma he faced in 1992: “I want to get the best computer for computational biophysics, and what that is, I have to determine myself.” So he hatched a reconnaissance mission. A very ingenious one. He would send a handful of students out across the country to computer companies and institutes and have them ask what is the best computer option to invest in. Schulten himself specifically did not go–he felt the computer companies would see his students as “innocent young people” and would not lie to innocents.
His students came back with an answer: Your best option is to connect a series of workstations together and make a cluster; don't go with any big computer options people are trying to sell you. Schulten had his answer! He would have to build another parallel computer, but it would be considerably less work than the homemade supercomputer he built in the 1980s, and from the reconnaissance missions, he knew it was unlikely to fail him.
It's a good thing Schulten didn't go with the Connection Machine. Right around the time Schulten was making his decision about how to spend his equipment money, the Wall Street Journal broke a story in summer of 1991 that some supercomputer companies were being unfairly subsidized by DARPA (Defense Advanced Research Projects Agency). DARPA had been founded in the United States in 1958 as an answer to the surprise launch of Sputnik by the Soviet Union. DARPA's mission was to expand the technology of the country beyond the immediate needs of the military. Essentially it functioned to make sure the surprise of Sputnik never occurred again–DARPA was a pure Cold War outgrowth.
It turns out DARPA was playing favorites with supercomputer companies, and the company that made the Connection Machine was a major recipient, to the tune of $55 million over the years the company was in business. When the story broke, it was a big embarrassment for the (elder) Bush administration. It also turns out that the company that made the Connection Machine was rife with mismanagement, paranoia, and ineptitude. (The full debacle of the Connection Machine is told in great detail by Gary Taubes in this article.) Without the DARPA gravy train, the company started losing money, and it became clear that their Connection Machine could not stand up to the performance of other competing supercomputers popular at the time. Klaus Schulten had escaped a Cold War minefield, by passing on the Connection Machine.
But Schulten found himself now out of favor with the supercomputer center at the university, for refusing their offer of the Connection Machine deal. But he never looked back. He had a vision, that the computer could be a very useful tool in biology, and that guided his decision to build a homemade cluster in 1993. “It is really some important instrumentation for biology that can be very useful,” says Schulten about the computer. “I wanted to make it useful and so I wanted to be sure that I really served my field and not my ego.” That basically summarizes the principle that guided him in his decision making. He wanted the best computer for computational biophysics.
But the computer cluster Schulten decided to invest in was only one part of the equation. His group needed to run molecular dynamics on this parallel cluster too. In 1994 Schulten's graduate students were working on porting existing molecular dynamics codes, which had been developed by previous group members in the late 1980s and early 1990s. But the students became so frustrated looking at this mass of indecipherable code. They proposed writing a new code from scratch, and this became a staple in the group from then on. The result was NAMD, a molecular dynamics code designed from scratch to be parallel and written in modern C and C++. (But a long story could be told about the scorn they met for their use of the software languages C and C++ in NAMD, rather than the venerable FORTRAN that every self-respecting computational scientist used at the time.) Between having their own parallel machines, and a new piece of software specifically tailored for parallel computing, the group was ready for the dawn of the new millennium. By then, Klaus Schulten's center had become expert in parallel computing.
Aquaporins, the Long Sought-for Water Channels
In 1985 Klaus Schulten witnessed the determination of the first structure of a membrane protein. At that time he set himself a goal of simulating a protein in a membrane. Some fifteen years later, at the turn of the century, his group was finally ready to meet the challenge–embed a protein in a membrane. The candidate he chose was a modest membrane protein. Its function was simply to let water in or out of a living cell. But the simplicity of this protein's functions belies the complicated storyline that would ensue for Schulten.

Aquaporin embedded in a membrane, with water permeating.
Water is essential for all living cells. For example, humans are about 70% water. But before 1992, scientists had no way to account for how some cell membranes seemed to allow permeation of significant amounts of water. Scientists knew that a small amount of diffusion through the lipid bilayer was possible, but several scientists felt that in cells like salivary glands, renal tubules, and red blood cells, permeation happened on a more massive scale than could be explained by modest diffusion through a membrane bilayer without the help of a special water channel. Thus began a search for a water channel.
Peter Agre calls his discovery of such a water channel “sheer blind luck.” In the mid 1980s Agre was studying the blood group antigen RH, and came across a smallish membrane protein that he viewed merely as a contaminant in his Rh preparations. It wasn't until some years later that he started wondering what this new membrane protein's function was. After conducting many different kinds of studies to learn all he could about the protein, he decided to talk to lots of other scientists, biochemists and physiologists, for clues. One of his friends he consulted in North Carolina on a drive home from a summer vacation in Florida said it sounded like the long sought-for water channel.
So Agre envisioned an experiment. Express the mystery protein into a system that is known for having low water permeability. The system he chose was frog eggs, which live quite stably in fresh water. And then, when he put into distilled water the eggs that he forced to acquire the mystery protein, they burst like popcorn! This meant that the mystery protein was allowing water permeation in droves in a system that normally lives happily in water. He had found the long sought-for water channel, which he coined an “aquaporin.” He published his results in April 1992.
By 2000, there were two published structures of aquaporins that caught Schulten's eyes. One was a human aquaporin, the other was from E. coli. It was an ideal system for Schulten, however, as this particular membrane protein had a simple function: conduct water. From a molecular dynamics point of view, this allowed Schulten the luxury of watching the membrane protein in the course of its function, which often wasn't visible via the many experimental techniques used to characterize the protein. Aquaporin simulations would then become a huge focus in Schulten's group for the next seven years, and lead to over fifteen publications on the topic.
Slings and Arrows
“These theoreticians! They play their games, and they agree with experiment. They don't tell you anything new. NO, not in this case.” Klaus Schulten grows animated when talking about molecular dynamics applied to aquaporins. But he reiterates a common dilemma for scientists doing computational biology. If your theoretical calculations agree with experiment, then they're not new; and if your theoretical results have no experimental corroboration, then they must be invalid. It's practically a no-win situation. But Schulten's aquaporin work really drove home the point for him that the computer can behave almost like a computational microscope. It can often capture a dynamic picture where experiment has a blind eye. In the aquaporin work that Schulten did, he is quite proud of one finding in particular–how these channels exclude protons from the outflow of water. But it was a hard-earned victory with many struggles.
By early 2001 Schulten already had slated for publication a slew of papers on the mechanism of conduction through aquaporins. It turns out that people found some aquaporin channels can conduct glycerols as well as water, and some of Schulten's work also focused on these so-called aquaglyceroporins. But Schulten realized he needed experimental collaborators to shore up his molecular dynamics findings. Schulten remarks that he often has a better chance of getting published if he's teamed up with an experimental group–one of the vagaries of life in computational biology. At this time, he had a visiting PhD student from Denmark, Morten Jensen working on the E. coli aquaglyceroporin. Jensen jointly with another postdoc Emad Tajkhorshid had been conducting research and running simulations on an aquaporin structure determined in 2000 by Robert Stroud's group in San Francisco. Schulten eventually convinced Stroud that the two groups should pair up.
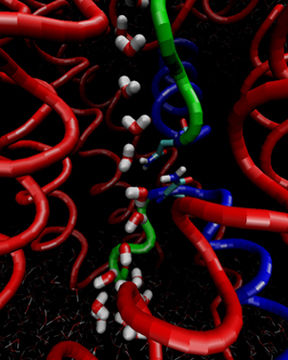
Water flipping its orientation as it flows through the aquaporin channel.
By that time Jensen and Tajkhorshid were running simulations on the new machine at the Pittsburgh Supercomputing Center, called the Terascale Computing System. At this point they really needed it, as the system they were studying–protein, membrane, water–was over 100,000 atoms. But with Schulten's software NAMD running on that terascale machine, timely calculations could be completed. This was considered a large system, for many researchers were only doing at most 10,000 atoms.
The simulations that Jensen and Tajkhorshid ran confirmed a hypothesis (from Yoshinori Fujiyoshi's group in 2000) that the orientation of the water molecules may have something to do with aquaporin's magical ability to prevent proton conduction. In essence, if there is a column of water molecules, protons can easily hydrogen-bond to a water, and then they hop from water molecule to water molecule, much like a hitchhiker jumping on a train hops from moving car to car. Basically, what simulation showed was that a water goes forward and into the channel with its oxygen atom facing downward, and then midway through the channel, the water molecule flips, with oxygen atoms facing backward, against the direction of movement as shown in the image. Bob Stroud suggested to run some simulations with certain key residue charges, which they suspected may be responsible for the “flip,” turned off. Indeed these simulations confirmed the residues responsible for the flip, and that pointed to why certain residues are conserved across aquaporins.
But what exact mechanism was responsible for the flip, and how did that keep protons from riding along with the water molecules through the channel. Jensen said there were protracted discussions about this. The collaborators eventually figured out that the symmetry of the protein channel contributed to the water flipping, and keeping the protons out. In the subfield of physics, electrostatics, an object with opposite charges at either end is called a dipole. Two dipoles together make a quadrapole. The inside of the aquaporin had just such a quadrapolar field due to its symmetry. The presence of two opposite dipoles effectively disrupts the protons from hopping down the chain of water, and hence the protons don't ever make it through. Since the inside of a cell is maintained at an almost constant voltage, the loss of protons would disrupt this “cellular battery” and render the cell dysfunctional. Incidentally, Schulten would later go on to find that the quadrapolar field also is at work in conduction of glycerol. Glycerol is a long linear molecule and to flip the molecule while it's in the channel would have about as much chance for success as a large cargo ship turning around 180 degrees in the narrowest confines of the Panama Canal. Instead, the glycerol molecule has three O-H groups, each one having its own dipole moment. The glycerol molecule, as it travels through the aquaporin, actually just rotates its arms (that contain the O-H groups) from up to down in the quadrapolar field, and so this is how glycerol rather than water moves through the channel, adjusting its dipole moment to the internal quadrapolar field.
With such careful attention to detail, a combined experimental and theoretical team, and the mechanism of proton exclusion explained, Schulten was floored when the editor of Science called him to explain why the journal was rejecting his paper. Apparently a referee slammed the aquaporin paper, gave it a literal thrashing. And, to rub salt in the wound, the Science editor revealed that another paper was about to come out that also was a molecular dynamics study of aquaporin. Schulten's team had been scooped!
Luckily, Schulten could bargain with the editor and Science gave him a shot to publish, but only if the teams did another test, add a mutant to determine the positions of the water. So, the experimental team added the mutant and then the theorists ran the calculations and got very nice agreement. But this delayed acceptance by four months. The teams eventually published their results in April 2002.
But that was not the end of the hardship for Schulten. Over a year later, an old friend of Schulten's published a paper claiming that the theoretical results about the proton exclusion that Schulten had elucidated were flat out wrong. “People have a suspicion whenever a theoretician does something, they think, 'Oh maybe it's not right,'” Schulten says about one of the perils of being a theoretical biologist. “And then, if another theoretician says it's wrong, it looks really bad.” That was exactly what happened to Schulten and aquaporin. “Our beautiful example got really spoiled,” he says. Schulten was at a loss for what to do. Basically he decided to do nothing, and hoped time will tell. Schulten's wait and see approach did eventually pay off. Today his aquaporin Science paper is a highly-cited publication.

The tetrameric nature of aquaporin.
For Morten Jensen, the visiting Danish student in Klaus Schulten's lab who did so many studies of aquaporins, the work he did had deep ramifications, perhaps for the good of society. He says while in Schulten's lab, they would talk about the technological feasibility of using aquaporins to filter water. When Jensen returned to Denmark, he actually went a step further than just imagining that aquaporins could provide a necessary technology–he co-founded a company to exploit aquaporins! He recruited many scientists to be part of the advisory board, and didn't stop there. “We got actually that first round of money that you need just to do the very first baby steps to get proof of concept, with relative ease,” Jensen recalls. The company, Aquaporin, has been going strong ever since, and is now working on ways to scale up the process for industrial uses like waste water treatment and possibly even desalination. While desalination currently requires lots of energy input to make potable water, the company in Denmark has developed a membrane full of the proteins that doesn't require high pressure to force water through the membrane–instead, osmosis is the key. With access to clean drinking water a global issue, and at least seven states in the United States suffering from severe drought, a technology like this has broad implications.
For Klaus Schulten, the aquaporin work was a real eye opener. He always suspected that the computer would be a good tool to use for membrane protein processes, and he finally had his proof. He saw three key areas where the computer was useful. First, large sizes were now attainable. Second, the disordered membrane environment is described well enough. And, lastly, the dynamic processes going on in membrane proteins, such as transport, could be resolved, and were especially useful when experiment was at a loss to yield such dynamic data. Despite his many setbacks, Schulten now had the green light to move forward and describe further membrane proteins, as we shall see below.
Entree to a New Class of Proteins: The Mechanosensitive Channels
Around the dawn of the new millennium, more structures of membrane proteins were being solved, and Schulten started to take advantage of this fact in his professional life. While his group was actively involved in aquaporin research, he decided to tackle another class of membrane proteins: the mechanosensitive channels. This would result in an object of study that is still boggling Schulten to the present, for it presented a conundrum. However, this same conundrum offers to Schulten a chance for discovery, and he is throwing at it everything he has. His love of discovery is, admittedly, the major impetus that drives him as a scientist.
Cells in prokaryotes and eukaryotes have evolved ways to respond to mechanical stress. For example, a bacterial cell likes to have the environment that surrounds it closely mimic the environment inside it. That is, the concentration of ions should be roughly the same inside and out. But what if there's a rainstorm, and suddenly the outside of the bacterial cell is mostly water, while the inside has water and solute? Water will rush into the cell, a process called osmosis. But then the cell will swell and, after a while, risk explosion because of the great pressure inside it. This is called osmotic shock. Enter the mechanosensitive channels in bacteria. There are three of them, and each of them kicks in as a sort of safety valve when the alert level is at different stages, much like airports were color coded with various alert levels in the years after 9/11. (To be clear, a fourth channel has been found in E. coli, called MscK, but only the canonical three channels will be discussed here.)
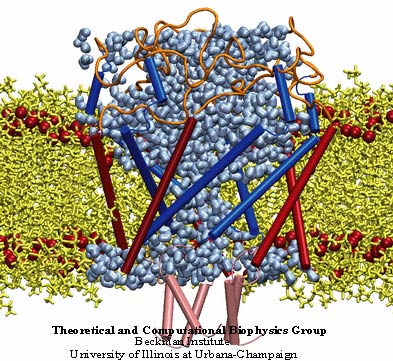
MscL or the mechanosensitive channel of large conductance.
When the lowest alert level in a cell is detected, the first mechanosensitive channel opens, and out go water and solute. If that doesn't take care of the problem, the next mechanosensitive channel opens, the one for medium alert level. And finally, a third one as a means of last resort will open to relieve the stress on the cell. But these channels open with cost to the cell. The solutes they release because of osmotic shock are precious to the cell's metabolic processes, so ejecting them is much like throwing off vital fuel supplies from a sinking boat.
While these mechanosensitive channels are central to cells undergoing osmotic stress, they are also implicated in touch and hearing. Furthermore, plant cells are thought to use these channels to detect gravity and distinguish the difference between up and down. In 1998 experimentalists determined a crystal structure of the mechanosensitive channel of last resort, and by 2000 Klaus Schulten was running simulations on it. This channel is called MscL, for the mechanosensitive channel of large conductance. The goal of the simulations in Schulten's group was to elucidate the gating mechanism, or what makes the channel open and close. These simulations, mostly done by graduate student Justin Gullingsrud, were novel for the fact that this was probably the first time a molecular dynamics simulation was run in the presence of applied surface tension. This would be Schulten's introduction to a class of channels whose characteristics were ripe for unearthing by computer simulation
The Allure of the Balloon
It seems like from the beginning, when Klaus Schulten saw the crystal structure of MscS, he was hooked–he couldn't believe how beautiful it was. The mechanosensitive channel of small conductance, or MscS, is located in the cell membrane of a bacterium, in fact spans it, forming a potential channel. Under normal condition the channel is actually closed, but under osmotic stress stretching the cell membrane, the channel becomes open. The opening kicks in when conditions are at a medium alert level; it is the second of three types of proteins to open. Ironically, MscS is a much larger protein than MscL, the channel of large conductance. In fact, the large size of MscS is one of the things that both fascinate and befuddle Schulten. A most compelling aspect of the large protein MscS is that a huge percentage of it (about 65%) resides OUTSIDE the cell membrane and in the cytoplasm. As shown in the picture, MscS looks much like a lantern, with the lower segment of it outside the lipid bilayer, and containing seven windows. This bottom part, or the cytoplasmic domain, has come to be known as “the balloon” in Schulten's group. When Schulten started studying MscS in 2003, he realized, as everyone was paying attention to the part of the protein that resided in the lipids (the transmembrane domain), that the nature and function of the balloon was a mystery. And he wanted to unlock it. Many seem to think the balloon is some kind of filter. Unfortunately, Schulten's first forays on MscS seemed to provide more questions than answers.
Since Schulten had already been studying mechanosensitive channels (in the form of MscL) when new graduate student Marcos Sotomayor joined the group around 2003, and a crystal structure of an MscS appeared for the first time, it seemed natural for Schulten to continue with this topic. Sotomayor says it was also a good problem for introducing him to the biophysics of proteins.
MscS, with the "balloon" the lower segment of the protein. Two windows are visible in the balloon.
Luckily, Sotomayor was very familiar already with molecular dynamics; he had written his own program to work on a physics-related topic: the phase transition in gases. He conducted this work while he was a physics student in Chile. Sotomayor relays that part of the reasons he picked the University of Illinois at Urbana-Champaign for graduate school was the broad range of areas covered by over sixty professors in the physics department. While he was shopping for advisors, Sotomayor saw a talk by Klaus Schulten on how birds use quantum mechanics to orient in the earth's magnetic field. This intrigued Sotomayor enough to approach Schulten. While Sotomayor was a bit worried that he didn't have a background in biophysics, Schulten was thrilled that he had molecular dynamics training. “I need people that know how to do simulations, and biology you will learn on the way,” Sotomayor remembers Schulten reassuring him.
When Sotomayor started his simulation of MscS, everyone, including himself, was under the impression that the 2002 crystal structure was that of an open state. This was because when the experimentalists crystallized MscS, they could see straight through an opening in the membrane protein. Basically, when water + ions are passing through the pore, this represents an open state; when ions cannot pass this represents a closed state. And a third category exists, that of in inactive state. The early simulations that Sotomayor and Schulten ran on MscS showed an asymmetric closure of the pore. Here the two scientists got their first clue that the 2002 crystal structure might not be one of the open state. Along with finding the asymmetric closure, they also found that the lipids interacted with the protein to a large degree, so the relationship between the membrane and protein was complex. In fact, Sotomayor says the complex lipid-membrane interaction “actually kind of determined this very dynamic motion of the transmembrane domain that led to the asymmetric closure.” In essence, Sotomayor and Schulten saw a big deformation of the lipid membrane around the protein. The also discovered that the pore in the transmembrane domain was somewhat hydrophobic, that is, water would permeate the pore, but only intermittently.
In the next stage of work about MscS, Sotomayor and Schulten decided to reproduce some experimental work. Since MscS is a channel through which ions flow, experimentalists had measured the conductances, and it was possible to reproduce these measurements in simulations. They teamed up with another scientist at the university there, Umberto Ravaioli, and his postdoc Trudy van der Straaten. What the scientists discovered was that the crystal structure they were using indeed was not that of an open state. So the theorists tried to generate possible open channels in their calculations. “And I think that was really important because that guided future experiments trying to search for a wider open channel,” explains Sotomayor.
Sotomayor agrees that these first two studies, which produced two papers, seemed to raise more questions than they answered. “It was an exploration right,” says Sotomayor about his first two publications, “and we were just looking at something new and then finding things that we didn't know exactly what they meant.” For one, these first two papers suggested that the 2002 crystal structure was either an inactive state, or a closed state, but the theorists were unsure which. At the same time it was not clear what was the structure or the conformation of the open channel. In Schulten's group, in work that was done by then postdoc Alek Aksimentiev, a method had been developed to look at the electrostatic potential across a pore. Sotomayor relays that when he calculated the electrostatic potential for MscS, it was very peculiar–another mystery. Also unanswered was exactly how the gating mechanism functioned, what were all the variables that contributed to going from an open to a closed state. And then there was the balloon. What was its function, and how did the seven windows on the side work? Did the balloon play a big role in gating?
There are many reasons why the protein MscS is so difficult for both experimentalists and computational scientists alike. It is a large protein, over a thousand amino acids in total. It has seven subunits. As Sotomayor mentions, with its large size it is challenging to pinpoint which sections are most relevant to the protein's function. And it operates in the cell on the millisecond time scale. In the current climate it is possible to simulate hundreds of nanoseconds. But to see the function of MscS unfolding, 1 millisecond translates to 1,000,000 nanoseconds! Back in 2003-2005 when Sotomayor was running simulations, he already was pushing the size limit with this 224,000 atom structure, and could only simulate a few nanoseconds,
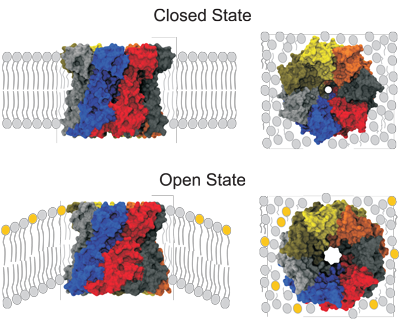
The different states of membrane protein MscS.
When Sotomayor and Schulten published their first two papers, they had a great suspicion that the crystal structure they used was probably not an open state, but they needed deeper analysis to probe this dilemma. In fact, this is a prime example in Schulten's career when the computer can help in determining what are the different states of membrane protein channels, for some of the states are not necessarily amenable to crystallization. But to make the best possible deductions, Schulten realized he would need to team up with an experimental group. Thus began a fruitful collaboration with friend Eduardo Perozo, an expert in molecular physiology and biological physics, who focuses his research on the dynamics of ion channels. Perozo's student, Valeria Vásquez, was working on MscS at the time, and so she and Sotomayor communicated frequently about what the other was doing. Sotomayor had to be sure the simulations he was running coincided with the experimental conditions Vásquez was creating in the lab. The two teams then set about the difficult job of deciphering the exact meaning of the crystal structure of MscS. In fact, in their first joint publication, they wrote:
Therefore, whether the crystal conformation represents an open, intermediate, inactive, or closed state, and how it goes from one state to another, are difficult questions, yet there exists an excellent chance to establish answers by combining experimental and computational approaches.
However, the world of the experimentalist and the world of the computational scientist are very different. As Klaus Schulten has noted, experimentalists often think what he does is just virtual reality. When Sotomayor visited the Perozo lab and talked to Vásquez and the other students, he sensed some tension. “I think the image that they had was that simulation people would just push a button and get the paper out,” Sotomayor notes, “while they were doing the hard work of the experiments, working every day on how to get their data.” But luckily that quickly changed for the better. Sotomayor thought it would be a great idea for Vásquez and her husband (who also was in Perozo's lab and became another collaborator) to attend a workshop Schulten's group was teaching on how to run molecular dynamics on NAMD. “I think at that point they actually realized that what we were doing is not Walt Disney Pictures, right, of a simple animation,” recalls Sotomayor. “It was real hard work behind the studies that we were doing.” Sotomayor also believes the experimentalists got a feel for what simulation could and couldn't do.
As it turns out, the Schulten-Perozo collaboration was a fruitful one. For example, while Vásquez measured the conductances through the channel of MscS, Sotomayor was successfully able to reproduce the measurements in simulation. The combined teams also could explain the gating behavior of MscS, which, in fact, yielded a publication in Science. But most importantly, the collaboration confirmed that the original crystal structure was not that of an open state, but rather, an inactive or closed state. Sotomayor and Schulten were able to incorporate experimental data from Perozo's lab and generate some possible models of an open state for MscS, which is a classic use of the computer in the elaboration of membrane protein states. In fact, Sotomayor believes the experimental data were really “a key to guide the search of open states.”
As it turns out, right about the time the collaborators published their 2008 Science paper on the gating mechanism of MscS, a paper also came out in Science of extreme interest to the Schulten-Perozo teams. An experimental group, led by Ian Booth and James Naismith, actually determined an MscS crystal structure with a pore much wider than the original crystal structure of 2002. Sotomayor says the theoretical predictions of the open state he and Schulten proposed, which were guided heavily by experiment, very much resemble the open state of the 2008 structure. But, as usual with MscS, all the details of the open state are still a matter of debate and some confusion.
A Methodological Development for an Enigma
“I felt, AH, there's a real secret there!” To the present, the mysteries of the MscS balloon are still challenging Schulten, but it seems like he actually enjoys this dilemma and generating theories to test. It is generally agreed upon that the balloon is some kind of sieve or filter. If MscS opens when a cell is swelling to the point of bursting, and just lets anything pass through to relieve the pressure, then lots of vital components will exit the cell and create an unfavorable situation inside. For example, the inside of a cell is usually at a negative voltage, which is a way for the cell to store energy to run processes. If important charged particles exit unencumbered through the balloon, the negative voltage a cell naturally maintains could be in jeopardy. So Schulten feels that the balloon is not just a passive filter, but that something more intricate is going on.
Right now, Schulten is in the early stages of a project to identify the function of the balloon, and he is quite excited about it. Currently, his graduate student Ivan Teo is undertaking studies on the MscS balloon. “What he is doing,” says Schulten about Teo's work, “is very interesting not only from an MscS point of view, but it is also very interesting from a methodological point of view.” Teo and Schulten are combining brainpower to uncover the secrets of the balloon, crafting a general method to apply to MscS, but which is general enough that it could be applied to other membrane processes. And their work is a prime example of how the computer can capture the myriad details of biology to yield discovery.
So far, Schulten and Teo have crafted a way to describe the ions moving in the area around the balloon in the cytoplasm. The pair has a long way to go to describe everything going on in the environment around and inside the balloon, but they are getting closer to honing in on a complete methodology to use. Schulten is hoping his tenacity over almost a decade will pay off. Only time will tell.
Translocons: The Causeways of Tenochtitlán
The beautiful ancient Aztec city of Tenochtitlán was built in the middle of a lake, and flourished during the 1400s. On examination of the archaeological evidence that has pieced together how it looked and functioned, it evinces remarkable likeness to a living cell. Tenochtitlán had a sacred area inside, replete with temples and a pyramid, and enclosed by a wall, much like the eukaryotic cell has compartmentalized sections enclosed by membranes. There were all sorts of specialized neighborhoods inside the island city-state, mimicking the societies inherent in a living cell.
Tenochtitlán, from a Diego Rivera mural at the Palacio Nacional de México. The middle of the image contains a causeway out of the city-state.
When James “JC” Gumbart started graduate school in Urbana-Champaign in 2003, he didn't know anything about biophysics, and assumed he would probably end up studying condensed matter physics, a strong suit for the university. “But then I TOOK a biophysics class actually in my first semester and was very intrigued by it,” Gumbart recalls. It was only a matter of time before Gumbart started working with Schulten, for Schulten's computational focus also resonated with Gumbart.
In the summer of 2004, when Gumbart joined the group, Schulten showed Gumbart a Nature paper about a protein-conduction channel, which Schulten thought was interesting and maybe could be a fruitful project. Probably to Schulten's delight, Gumbart indeed turned it into his focus, took lots of initiative, and would go on to produce an impressive body of work.
In only a matter of three years, Gumbart and Schulten produced three papers about various aspects of translocons that they unveiled through simulation. For example, in Gumbart's first paper, they studied a protein-conducting channel whose crystal structure had just come out, and about which little was known. What was clear was that this channel had a dual function: it either allowed complete passage of a protein to the other side of the membrane, or it diverted it to its final home inside the membrane. The pair of scientists in Illinois decided to focus on the former function of the translocon. And they actually uncovered a dynamic picture of what was going on. Gumbart pulled a short segment of amino acids through and saw a plug in the channel move to allow passage of the segment. While Gumbart admits this was a rather simplistic approach, it highlights the role of the computer for viewing dynamic processes of membrane proteins and their function. It also highlights that Gumbart learned a great deal (computationally and science-wise) in his early studies of protein-conducting channels, knowledge that would serve him well for the next odyssey about to begin.

JC Gumbart and Klaus Schulten and a ribosome attached to a protein-conducting channel.
Around the time Schulten was examining translocons, his research interests suddenly expanded to include a fascinating and massive molecular machine that has captivated him ever since. That molecular machine in question is the ribosome, which is the biological factory that assembles the cell's proteins. In 2004 an experimentalist approached Schulten at a conference and requested Schulten's help with a dilemma involving structures of ribosomes generated by two different kinds of microscopes. One type of microscope involves X-ray crystallography, which generates a structure of high resolution; the other type is electron microscopy, which generates structures of low resolution. This experimentalist (Joachim Frank) had been generating structures of ribosomes via cryo-electron microscopy and wanted help incorporating high-resolution data (from X-ray crystallography generated by others) into his maps from the electron microscope. By analogy, consider two photos, one of a bird sitting on a wire, and one of a bird in flight. The bird-in-flight image captures the bird in the act of flying (its function), but is low resolution. The bird on the wire snapshot, imagine however, can capture details such as every last feather. If somehow one could use all the fine details of the still bird and apply them to the bird in flight, one might be able to explain the bird's function–flying. Schulten's mission, therefore, was to take data from X-ray crystallography and fit it into cryo-electron microscopy maps; these electron microscopy maps were actually able to capture the ribosome in the midst of performing its function, unlike the false environments captured by X-ray microscopy. How much better you could explain ribosome function if you could imbue with fine detail that molecular machine in action.
Schulten knew in principle he could take this on. His group had been working on large systems for decades, he had in-house software products as tools, and had a coterie of students gifted in the computational arts. The result was a method, called MDFF or Molecular Dynamics Flexible Fitting, and it made its debut in 2008. More details about the genesis of MDFF can be found in this history. Suffice it to say, it brought to Schulten the riveting world of the ribosome.
But how does the ribosome relate to a protein-conducting channel? Ribosomes make proteins, as noted, and the newly created protein (called a nascent protein) exits its tunnel and then has to go somewhere. One option is that the nascent protein may go live in the cell's cytosol. Or its fate may be a voyage through the membrane to the outside. Yet a third option is the nascent protein might be destined for life inside a membrane. For these latter two, the protein-conducting channel steps in and guides the nascent protein to its target. In 2007 Schulten and Gumbart became aware of a group that had published a cryo-electron microscopy map of the ribosome in complex with its protein-conducting channel. (That protein-conducting channel is known as SecY) Here was an intriguing system–two macromolecules together caught on camera, each one influencing the other. More details about ribosome function could be unearthed if it was studied in tandem with its protein-conducting channel, which influenced it. And Schulten and Gumbart realized they could generate high-resolution structures of each part in the duo, to eventually fit into the electron microscopy map. It seemed like the perfect job for MDFF. Gumbart even went to Boston to get the electron microscopy map from the experimentalists who generated the complex.
Indeed, this project that Gumbart was working on was pivotal in many ways. It was one of the first applications of the new method MDFF in Schulten's group. Gumbart relates that the group was still ironing out the method while he was working on this complex of ribosome plus channel in January of 2008, but that it was a very exciting time, especially rewarding to see how the structures almost fit magically into the electron density. MDFF produced a complex of 2.7 million atoms, which included ribosome, protein-conducting channel, membrane and water, the largest to that date in Schulten's group. And Gumbart and Schulten could actually see how the ribosome induced a slight destabilization in the plug of the channel. It was a very successful project, and was just a prelude of what was to come.
As a result of the method MDFF, Schulten gained a new experimental collaborator in Munich, a biochemist named Roland Beckmann, who captured exciting systems with his electron microscope. Beckmann was already working on electron microscopy of a ribosome connected to a protein-conducting channel when he and Schulten joined forces. In fact, an early project between Schulten and Beckmann earned the two teams a Science publication, in which MDFF was used by Gumbart on a map Beckmann had made of a mammalian channel with a bound ribosome.
Klaus Schulten will always remember the first time he visited Beckmann in Munich. It was July 2008, and during a meeting in Beckmann's office, Beckmann took Schulten outside to show Schulten a poster on the wall of an electron microscopy image. And Schulten nearly fainted in delight when he saw a particular object in the poster. Without Beckmann describing anything, Schulten immediately recognized there in the map a nanodisc, an object invented at the University of Illinois. Schulten had helped the inventor of the nanodisc visualize it, so it was something Schulten worked on earlier in his career. Basically, the creator of the nanodisc, Steve Sligar, wanted to fabricate a nano-sized chunk of membrane to hold membrane proteins, for as already noted, working with membrane proteins outside the membrane presented experimentalists with many problems. So Sligar gave to the world a recipe for creating a bundle of lipids held together with two scaffold proteins, an instant patch of membrane to hold your membrane protein!
Ribosome and nanodisc, a challenging but captivating system.
The innovation of what Roland Beckmann did was to take a ribosome, that had a nascent protein not quite fully exited from the ribosome tunnel, and introduce a nanodisc into the system–then the nascent protein was snaking into a protein-conducting channel in the nanodisc. In essence, Beckmann caught a snapshot of ribosome with nascent protein entering a channel in the environment of a membrane. And Klaus Schulten recognized the nanodisc immediately. Which is saying something, because when Schulten helped Sligar visualize the nanodisc back in 2005, Schulten only got indirect information about its shape. In Schulten's mind the nanodisc looked an awful lot like what he saw on Beckmann's wall that summer day in 2008, so this was a confirmation of the image Schulten presented to the scientific community.
Beckmann wanted Schulten's expertise in MDFF to make a high-resolution structure of the ribosome-nanodisc system. It seemed natural for Gumbart to work on this project, as it involved a protein-conducting channel. This was a challenging project, however, according to both Schulten and Gumbart. Gumbart was just learning how to make sense of electron microscopy maps. “So I had to stare at it for hours and hours before I could really start to, in my mind, see the protein coming out of it. When I say protein coming out of it, what I mean is, actually seeing something that represents a protein,” recounts Gumbart. But luckily he had both student pioneers of MDFF on hand to help him make sense of the maps.
Gumbart and Schulten were eventually able to make sense of the super complicated but groundbreaking system Roland Beckmann had captured with the electron microscope. For Gumbart, it felt like the culmination of all his PhD work up to that point. One of the goals he and Schulten had put forward for themselves was to see how membrane proteins get made. “So for the first time we were actually seeing this in action,” Gumbart recalls. For Schulten, this project with the ribosome and nanodisc was a highpoint in his career. He feels this project highlights how computing (via MDFF) can become a key partner to experiment. But most of all, he never expected to revisit the nanodisc again, and he never dreamed it would be for something so innovative.
YidC: A Continuation of Protein-Conducting Channels
“He said that the odds are the project is going to fail. It's ninety percent odds this project is going to fail.” Despite this warning by Klaus Schulten, postdoc Abhishek Singharoy decided to give it his full attention. In summer of 2013 Schulten was visiting Munich, and came back with some new projects, one of them with Roland Beckmann's group. Despite the dismal odds, Schulten probably was interested in the topic because it was a continuation of his many years work on protein-conducting channels. As if ribosome, nascent protein, and translocon weren't enough, it was known that an additional membrane protein, called YidC, sometimes assists the canonical protein-conducting channel. This was tantalizing to Schulten, who is always thinking about systems with multiple proteins. But Schulten was fully aware of the odds of success when he presented Singharoy with the challenge: the experimentalists need help from computational people finalizing the structure of YidC
You see, no crystal structure existed for YidC. It didn't look like one would be forthcoming for years or decades, either. But Roland Beckmann thought the structure could be constructed even without X-ray diffraction. The experimentalists in Beckmann's lab, when they couldn't get a starting structure for YidC, turned to a computational group in their home university (The Ludwig Maximilian University of Munich) known for its software tools dedicated to predicting protein structure from sequence. From this starting structure, the task next fell to Singharoy to work hand in hand with Beckmann's group on finessing the structure. Singharoy would use all the tools in the arsenal of Klaus Schulten's group, and the Beckmann experimentalists, primarily graduate student Stephan Wickles, would focus on electron microscopy and biochemistry. It was a perfect example of computer working in tandem with experiment. And it was a journey filled with drama.
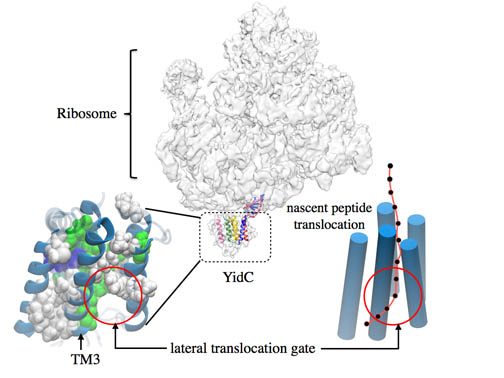
YidC, which facilitates lateral insertion of a protein into a membrane.
The computational group in Munich (led by Johannes Söding) derived a starting structure based on their software tools that employ bioinformatics. Not only were they able to predict the sheets and helices of YidC, but they also used evolutionary covariance analysis to predict, say, how close one helix may be to another. Now with a starting structure, enter Singharoy and Wickles.
With an “input” structure on hand, Singharoy placed the protein YidC in a membrane and started running molecular dynamics simulations. “But it was not normal molecular dynamics simulations,” Singharoy recounts. “It was MD that was constrained by the covariance measurements that we already had.” After the structure was run with restraints, then those restraints were removed and molecular dynamics was run again to equilibrate and to see the relaxed structure. Then it was time to send to experimental collaborator, who checks for which interactions make that relaxed structure stable. To do this biochemistry, Wickles would mutate the residues and thereby remove the interactions proposed to be the stabilizing ones. Then the process starts all over–molecular dynamics with and without restraints, then biochemistry experiments. Singharoy recounts they went through many iterations this way. All with the goal of finding the most stable structure.
After these tedious iterations between experimentalist and computational modeler, Wickles figured out a way to check that Singharoy's structure was a true energetic minimum, not some local minimum that YidC ended up at because the simulation time was not long enough. Wickles did a chemical cross-linking experiment. Basically, if two residues are close, by adding sulfur groups to the two residues in question, they can form a disulfide bond and this can be detected, thus proving that the two residues in question are nearby. What Wickles did, to test Singharoy's structure, was to insert a nascent protein into YidC and cross link a residue from each. And lo and behold, they were linked! It was the strongest test yet that indicated the many iterations had paid off and the structure was a meaningful one.
And then, in the midst of all their hard work, Wickles learned the unthinkable: another group had scooped them! A crystal structure was about to be published of a bacterial YidC. Wickles was at a Gordon Research Conference, and had a poster about his YidC work, when he saw the crystal structure on another poster. If there was a silver lining, it was that Wickles's noticed his modeled structure looked very much like the crystal structure he was seeing at the conference.
This set off a wave of frantic preparations to try and get a publication out as soon as possible. The three teams were about ready to submit a paper anyway when Wickles saw the crystal structure at the conference. Finally, after much trial and tribulation, the paper was accepted in July 2014, and came out only three months after the crystal structure was published by a team from Japan. The model-based structure that the triad of teams published was not the only significant result of the paper. Through the extensive molecular dynamics that Singharoy ran, he started to uncover some of the details about how YidC actually works, for he could see it dynamically in his simulations. For one, the simulations showed that the bottom of YidC forms a seal. This makes sense, as it is known YidC is an insertase, that is, it takes a nascent chain from the ribosome and guides it sideways to its permanent home in the membrane. No kind of channel opens up at the bottom. Instead the newly formed protein goes through a gate in the side of YidC. Simulations showed that one of the helices that formed the gate acts very much like a paddle, a flexible paddle, to allow nascent protein to snake into the membrane.
All in all, this project looked very risky from the beginning, but the tenacity of the investigators prevailed. They even fought through the drama of finding out that another group had produced a structure, and managed to get their paper published despite a dire situation. Singharoy indicates that the future holds some further refinement of the modeled structure based on the crystal structure, although the two structures are from different bacteria. And finally, the teams want to combine YidC with the ribosome, an endeavor that is sure to bring more excitement and interesting research, and probably will guarantee more hand in hand collaboration between computational scientist and experimental scientist.
BAR domains and Membrane Sculpting
New graduate student Ying Yin vividly remembers the day in 2006 Klaus Schulten called her into his office and produced a rumpled piece of paper from his pocket. He handed her the paper and told her, “We should work on this.” On the paper she only saw three letters, B…A…R. BAR. Not really much to go on. But over the next few years, those three letters would lead to a project that drew on every tool in the Schulten group arsenal. And a constellation of factors would come together to produce a beautiful application of molecular dynamics. This project, actually, is a consummate example of how computational biology can yield insights on dynamic processes associated with membrane proteins. Below we shall see how a cellular activity, membrane sculpting, was elucidated in its entirety from beginning to end.
Just what is membrane sculpting? A eukaryotic cell is a complicated place with many compartments walled off by membranes. These membranes come in many different kinds of shapes–flat, curved, wavy, or some very regular. There are also vesicles in a cell; for example, in the Golgi apparatus (the amazon.com of the cell) proteins are packaged up inside membranes (one type of vesicle) and sent to their final home, either in the cytoplasm, or perhaps to be released to the outside of the cell. The point is, there are many membrane systems within a cell, and each of those systems has a unique shape. As Klaus Schulten says about these myriad membrane systems in cells, it's all “FORMS, FORMS, FORMS!” But what exactly is responsible for shaping so many different forms? Often, proteins are used to “sculpt” the membrane into its final shape.
When Klaus Schulten gave Ying Yin that piece of paper with three magical letters, it actually referred to a family of proteins called BAR domains. These proteins are known to bend membranes (during endocytosis and exocytosis). There are actually three members of the family (N-BAR, F-BAR, I-BAR), and Yin's focus shortly became N-BAR proteins, for which there was a structure. As she started investigating the protein, she became more and more interested and more and more convinced it was a topic worthy of study. And Yin started discussing N-BAR domains with her fiancé, Anton Arkhipov, also a graduate student in Schulten's group. From these discussions, in the great lattice of coincidence, the project got a big boost. The couple realized they could collaborate and thereby simulate what before seemed impossible. Even Schulten never expected such simulations were feasible. What did the couple hit on?
Comparison of all-atom representation versus coarse graining on a single BAR domain protein.
Yin and Arkhipov actually met during their very first days of graduate school at the University of Illinois, in August 2004. They crossed paths at a qualifying exam, and realized they were both new students in the Physics department, Yin coming from China and Arkhipov from Russia. Arkhipov, fascinated by neuroscience, soon joined Schulten's group, for he had learned about Schulten's past work in neuroscience and then became fascinated with the idea of computational biophysics that Schulten currently worked on. Yin started working in a high-energy physics lab but realized she wanted to do something related to life, and so Arkhipov suggested she join Schulten's group.
Arkhipov had many diverse projects in Schulten's group, but one of the things he was doing, along with graduate student Peter Freddolino was implementing coarse-grained molecular dynamics into NAMD. In this method, a group of atoms is represented as a “bead.” For example, one bead may represent 10 atoms. While this may mean some loss of fine atomic detail, it essentially equates to being able to run molecular dynamics on extremely large systems and for extremely long times. Arkhipov was doing two kinds of coarse-grained molecular dynamics, one being residue-based and the other shape-based. The former has single-residue resolution (about 10 atoms per bead) and the latter uses a group of beads to represent a whole protein (about 150 atoms per bead). A comparison of all-atom representation versus shape-based coarse-graining is shown for clarity in an image of a typical BAR domain protein.
Arkhipov actually used coarse-grained molecular dynamics on virus capsids as an application of the method. Also at this time, graduate student Amy Shih used coarse-grained molecular dynamics for the nanodisc system that was mentioned above. “As Ying was working on BAR domains, we discussed it of course, just out of curiosity,” Arkhipov says. “And we figured out together that this could be a good application of this coarse-grained project.” The couple relates that Schulten was very supportive of their proposed collaboration. Schulten, in fact, has been collaborating with his wife, a chemist, for some forty years and understands both the rewards and potential professional perils of husband-wife collaborations in science. Yin and Arkhipov married in 2008.
In the initial work that Yin and Arkhipov did, they put one BAR domain protein, which is shaped more or less like a banana, atop a patch of membrane and watched as molecular dynamics showed the membrane near the protein bending to fit the curve of the crescent-shaped BAR protein. And then they tried something completely different. They put six BAR proteins down in a line on a patch of membrane (see image). The only reason they could even attempt such a feat was because of the new NAMD feature, coarse-graining. Without the simplified representation, it would have amounted to a few million atoms and they could never have simulated for a long enough time to see if the six proteins bent the membrane. When the six BAR proteins produced a wavy shape, they tried laying down six proteins in a lattice formation (see image). Voila! They had hit on a configuration that produced beautiful curvature. It was a lattice of BAR proteins.
The membrane curvature produced by multiple BAR domains. On the right is an example of a lattice of BAR domains.
Schulten relates that when they tried to publish their results with a lattice of BAR domains, they had trouble. Referees pointed out that in the cell, these high concentrations of BAR proteins are never found, so the simulations seemed unrealistic. “But then people succeeded to take electron micrographs of the membranes together with the BAR domains,” notes Schulten. “And then they saw they formed exactly the lattice that we talked about!” So now it seemed likely that many BAR domains worked together to sculpt membranes. Shortly thereafter their paper was accepted.
As a grand finale, Schulten, Yin and Arkhipov decided to really test the new method, coarse-grained molecular dynamics, and see if they could form a tube out of a flat patch of membrane. “Psychologically, and just as a demonstration of the power of the method, nothing can be better than actually showing the formation of a full tube,” notes Arkhipov. So Yin put down a lattice of N-BAR proteins on a 200 square nanometer membrane patch, submitted her job to the supercomputer, and waited. In fact, she waited almost 200 days, the good part of a year. However, Yin recounts that she checked progress several times a day, and often saw the same thing day after day–the tube wasn't closing. Schulten told her to be patient. Then one day when she checked, a perfectly closed tube had formed! All three scientists were thrilled. Coarse-grained molecular dynamics could yield a thorough look at membrane sculpting in action. As Arkhipov sums up, “I was very happy to see that, because it's easy to invent a method, but it's not easy to make it useful.”
Schulten has continued his work with BAR domains to the present. Currently his graduate student, Hang Yu, works on F-BAR domains. Inspired by a 2008 cryo-electron microscopy image of a lattice of F-BAR domains on a tubule, Yu and Schulten set about answering a series of questions for themselves: How does an F-BAR domain work in a cell, does it sculpt a membrane like N-BAR proteins? Why does a cell use that specific concentration of F-BAR domains?
So Yu set about to look at how an F-BAR domain curves a piece of membrane. When he ran his simulation, nothing happened, the membrane just stayed flat. He ran several simulations and actually saw nothing. But he was using comparable times that his predecessors, Yin and Arkhipov, used. Yu finally decided to let the simulation run for an extremely long time, and lo and behold, he finally saw the F-BAR domain curve the membrane. What Yu discovered was that F-BAR domains are less rigid than N-BAR domains, and F-BAR domains also produced less curvature.
A lattice of BAR domains sculpts a membrane into a tube.
Yu then tried to find what optimal density of F-BAR domain proteins produce the highest curvature. On a 1000 square nanometer piece of membrane he laid down either 5, 8, 10, or 16 dimers. He saw that the 10 dimer arrangement produced the best curvature on the flat membrane. Now Yu was armed with the specific lattice that produces the highest curvature. He was ready to see if F-BAR domains would produce a tubule from a flat piece of membrane. Yu reports that he and Schulten were thrilled when they saw a tube completely form.
In all of Schulten's work on BAR domains, he argues that what actually makes the membrane curve is called the scaffolding mechanism. In this mechanism, there are positively charged residues on the underside of the banana-shaped BAR domain that attract negative lipid heads. This attraction draws the lipids to the BAR domain and in this way curves the membrane. But the jury is still out on the sculpting mechanism, for some people favor another method, called the insertion mechanism. In this mechanism, residues of the BAR domain are actually inserted into the membrane and pinch it, causing it to curve. While Schulten argues for the scaffolding mechanism in his work on N-BAR domains and F-BAR domains, he still gets grief from people proposing the insertion mechanism. Schulten does acknowledge that at times in a cell, some proteins might use the insertion mechanism to curve membranes. “Because if you look in the cell,” clarifies Schulten, “you see so many different shapes. I mean so many that you would never think they are all done with one trick.” But he is very confident in the scaffolding mechanism for BAR domains.
Insights on the Nervous System
When Klaus Schulten heard around 2005 that a structure had been solved for a voltage-gated potassium channel, he was intrigued, for this indirectly linked to his years of studying the brain. While potassium channels were a trendy topic, Schulten was motivated instead by his long-held fascination with brain research. And potassium channels are key to nervous system function.
The nervous system in animals is the primary way animals react to the outside world, with the central nervous system, the brain and spinal cord, being the largest component of the nervous system as a whole. Nerve cells detect stimuli and then pass this information from neuron to neuron, down the long axons. Electricity, or the flow of charges, is key to this transmission from one neuron to the next, but unlike a building with electrons flowing in its wiring, the brain's electricity is carried by ions, namely potassium and sodium.
But how does a nerve cell fire? In essence, the inside of a cell is constantly maintained at a small negative voltage (this functions as a battery of sorts). In human cells, the inside contains many potassium ions and the fluid bathing the outside contains many sodium ions. When a stimulus (sound, touch, light, etc.) reaches the cells in a sensory organ, it is detected and converted into an electrical signal that reaches, in a single step or after multiple steps, neurons heading towards the brain. In these neurons sodium channels in the membrane of the cell open and sodium ions flow inward. Eventually, when the voltage reaches a certain value, this causes a plethora of more sodium channels to open and eventually a threshold (positive) voltage is reached inside the cell. At this point, about a millisecond later, an action potential occurs–a somewhat large positive voltage is reached. Sodium channels now close. Next, potassium channels open and potassium ions leave the cell, returning the cell to a more negative voltage than where it initially started. Hence, the cell overshoots its mark. A pump then restores the cell to its correct small negative resting potential by pumping potassium and sodium in and out, respectively. So this transport of ions occurs across a membrane, and gets passed down the membrane of the axon of the nerve cell to transmit a signal. But this electrochemical behavior is all based on sodium channels and potassium channels. With the solution of a potassium channel on hand, Klaus Schulten realized he could examine a key element of a neuron from a physics point of view.
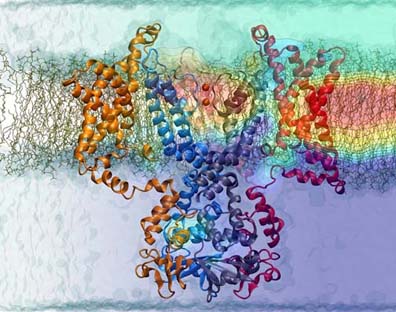
Voltage-Gated potassium channel embedded in a membrane.
Around the same time Schulten learned of the new structure of the voltage-gated potassium channel, he also had a new student, Fatemeh Khalili-Araghi, who began studying the topic. Initially interested in high-energy physics when she entered graduate school, she soon realized the physics in Schulten's group was just as captivating as that in string theory. She immediately started investigating the potassium channel. Scientists knew it was a kind of channel that opens and closes (called gating) based on changes in voltage. “When I started reading more, I realized we didn't know how they sense voltage. We didn't know what happens inside,” says Khalili-Araghi. “I mean we basically knew nothing.” It was Khalili-Araghi's job to learn more and elucidate the mechanism, but it would prove to be a major challenge.
She and Schulten first set out to understand the permeation of potassium ions through the pore. There is stretch of five amino acids in the protein that form the narrowest segment of the channel, and which keep out everything but potassium, hence it is known as the selectivity filter. Much had been postulated about how the ions pass through the selectivity filter, one idea being that potassium doesn't flow continuously. When Khalili-Araghi started simulating potassium flowing through the channel, it wouldn't conduct. This was odd, because the crystal structure that had been solved was purported to be in an open state. She went through a myriad of tries before she realized that an “inactivation” of the filter was occurring–it was something that happened spontaneously and prevented conduction. What she eventually realized was that some carbonate groups in the filter were responsible for the non-conduction, and after many tries she constrained them just right to see conduction. And finally she and Schulten confirmed that permeation of potassium ions occurred through a knock-on mechanism; there are two potassium ions in the channel, and then a third comes in, it stays there but pushes the bottom potassium down through the rest of the channel. While this confirmation netted a publication in Biophysical Letters, Khalili-Araghi says the tribulations of simulating the conduction were not even the hardest part of the project.
Around the time that she was investigating the potassium channels, Khalili-Araghi was at a Biophysical Society meeting and she struck up a conversation with a scientist from Benoit Roux's lab in Chicago. Roux is a biophysicist who was studying potassium channels as well. Khalili-Araghi learned that a collaborator of Roux's, Vladimir Yarov, had built a model of a closed structure of the voltage-gated potassium channel. This soon became valuable for Khalili-Araghi, and she and Schulten would soon begin a collaboration with Roux's lab. While membrane proteins are traditionally difficult to crystallize, it is even more difficult to crystallize an ion channel in a state that only exists upon application of a specific voltage to hold it closed–for that state must happen inside a membrane. But the closed state model from Yarov was not like a pristine crystallographic structure. Khalili-Araghi had to spend extensive time refining the structure until she had a stable closed state.
But her work was not over. “After that, we had a closed state and an open state, and we had to verify them somehow, that the closed state is a real closed state,” she summarizes. What ensued next was some serious computing–calculation of the gating charges. When the channel goes from the open to the closed state, there are some charged residues that move, and these movements can be quantified in experiments. While Khalili-Araghi could exactly reproduce those experimental measurements, to say that the simulations required massive computer time is almost an understatement. “At that point, I think the amount of computer time that we used for this project was probably similar to all of the other computer time that people used in Klaus's group for all of their other projects combined at that time,” says Khalili-Araghi. But the efforts were well worth it, for they got a very close match to experimental gating charges. Khalili-Araghi did many more calculations to verify and improve the closed state. In the end, they found a structure that had a narrow constriction region with negatively charged residues. “So everything was falling into its place, again pointing out that our closed state is probably something similar to the real closed state found in the cell membrane of a neuron,” she contends. “It's probably the closest we can get, I mean until we see that from crystallography or any other method.”
Tetrameric structure of the potassium channel.
So what did the long simulations of the neuronal potassium channel reveal at the end? They actually answered a crucial question regarding a fundamental property of neural signaling. This signaling comes about through a voltage dependence of the conductivity of sodium and potassium ions across the neural cell membrane in the atonal region of nerve cells. In the case of both sodium and potassium channels, the conductivity is realized by a central pore that discriminates with high specificity for either sodium or potassium ions. This pore is surrounded by four so-called voltage gates that control the conductivity of the central pore, for the purpose of sensing the voltage across the neural membrane. As ions are conducted the voltage actually changes and accordingly the conductivity changes then. As a result the ion conductivity is non-linear with respect to the voltage and the non-linearity shapes the nerve signals.
This mathematically interesting control of nerve signaling was discovered in the 1950s by Hodgkin and Huxley, long before any molecular detail was known about the ion channels in the neural membrane. Hodgkin and Huxley were awarded the 1963 Nobel Prize in Physiology or Medicine. Most amazingly, the mathematical form of the Hodgkin-Huxley equations is reminiscent of the recently discovered structure of the ion channels, indicating clearly that the ion channels should have four voltage gates. Still, a mystery, until Schulten became fascinated with potassium channels, was how the voltage gates manage to sense the weak voltage change that occurs across the neural membrane. Khalili-Araghi and Schulten's simulations showed in the case of the potassium channel that the voltage change is localized over a surprisingly short distance in the gates such that even though the change is small, the ensuing force (which is voltage change times ion charge / distance) is sufficiently high to produce a strong effect. The physics involves a redistribution of highly polarizable water molecules and a change of an ordinary protein alpha helix into a so-called 3-10 helix. Evolution discovered here an electronic nanodevice like a transistor that permits neural signals and thereby neural computation. Presently, the Schulten group is extending the studies of Khalili-Araghi to the sodium channel, which is more sophisticated in its function than even the potassium channel. Schulten is confident he will soon complete the bridge between structure and dynamics of the neural ion channels on the one hand and the mathematical phenomenology of Hodgkin and Huxley on the other hand.
The Grand Finale: A Series of Interlocking Processes
We are such stuff as dreams are made of…
––Shakespeare, The Tempest
Two of Klaus Schulten's dreams have defined his focus over the last forty-five years in his scientific life. As a young boy he had a fascination with ATP, and he never forgot this early allure with the universal currency of the living cell. In a similar vein, he wanted to study a key membrane protein, the reaction center, in order to explain photosynthesis, and his outrageous plan to build his own supercomputer in the 1980s steered him on the path towards becoming a computational biologist. So, over four decades later, is Schulten satisfied with his own personal understanding of either ATP or photosynthesis? He has gotten very close. And to represent the culmination of a life's calling, he has found it necessary to illuminate the series of processes in photosynthesis that begin with absorption of sunlight and end with production of ATP. “And the point is that what I wanted to do scientifically, I only could do through a movie,” summarizes Schulten. But this is no Hollywood production. Every single atom is correctly accounted for, and all the processes have been diligently demystified over the years by many, many scientists, including various researchers in Schulten's group. There are no special effects, just pure science. This movie, described below, also touches on another dream of Schulten's, his desire to describe how multiple proteins work together in a cell, forming societies of biological organization, which is the essence of making a living cell living. And this movie is based on a membrane system, the chromatophore of purple bacteria.
The spherical chromatophore, with its five different types of proteins in various colors. It functions in a series of interlocking processes.
The chromatophore is basically a self-contained unit in a purple bacterium that acts like a biological machine, making ATP from sunlight. It is actually called a pseudo-organelle, since it is found in prokaryotes. Basically, it carries out an autonomous function on its own, and while it only operates in the context of a cell, it is of such a size that it is now possible to simulate this “organelle” in its entirety, and thereby it brings Schulten closer to the goal of studying a whole cell. While there are many different shapes of chromatophores, the one featured in the movie is spherical, about 60 nanometers in diameter. It has over 100 proteins imbedded in its spherical membrane. Within the chromatophore, a series of processes take place between the proteins, which essentially interlock with each other. “They work like a Swiss watch in a way, with the little wheels, one driving the other and the other,” clarifies Schulten. “And it is this clockwork of processes that is really making the system do what it's doing.” While scientists over the decade have looked at individual processes performed by individual proteins, Schulten's goal with the movie was to show the sequence of processes that lead to the creation of ATP from sunlight, and illustrate the interdigitation of all the processes.
But how did Schulten get from a fascination with the reaction center (one of the five types of proteins embedded in the chromatophore) to an understanding of all the protein types that comprise the chromatophore? As soon as the reaction center structure was solved in 1985 by Hartmut Michel, Schulten immediately started running calculations on this membrane protein that is at the heart of photosynthesis. And then he took on a risky project. One night in Urbana during 1995 at a party in honor of Schulten's guest speaker Hartmut Michel, Schulten and Michel hatched a bold plan to determine the structure of a light harvesting protein. While the reaction center is critical to photosynthesis, there are a series of light harvesting proteins (shown in red and green in the image) that actually funnel energy to the reaction center, where the energy is then converted to electron transfer. The light harvesting proteins basically act like accessory antennae to absorb more available light for use by the chromatophore. Schulten's pact with Hartmut Michel was actually crazy, in hindsight, because Schulten was not an expert in structural biology, and Hartmut Michel's crystallographic data was missing a key piece of information (the phase angles). But Schulten, with intrepid postdoc, Xiche Hu, actually used computation to eventually figure out the structure of the protein called light harvesting complex II. So in this way, Schulten started to expand his knowledge of the protein components of the chromatophore.
As an aside, the solution of light harvesting complex II also led to what Schulten calls his biggest success in the field of membrane protein studies that he has worked on since the early 1990s. “And once we had the structure of the protein, then we immediately realized its value for biology, how it functions,” says Schulten about the light harvesting protein. At that point, Schulten took off the cap of structural biologist and put on the cap of quantum physicist. With an outstanding duo of graduate students, Thorsten Ritz and Ana Damjanović, as well as Xiche Hu, the team unraveled how the carotenoids and chlorophylls that make up light-harvesting complex II function as a unit and exploit quantum physics to accomplish efficient photon absorption and transfer of energy to the reaction center. On one side, Schulten was a biophysicist who dealt with protein simulations. “On the other side, I was a theoretical physicist who knew quantum physics well and so at that moment,” Schulten says of solution of the light harvesting structure, “suddenly my other half of the brain got into gear. And that was I think the most wonderful thing that happened to me.”
So initially Schulten was captivated by the reaction center, and then in the mid 1990s he started working on the two types of accessory proteins that funnel energy to the reaction center. Also, around 2005, Schulten started an ambitious project with postdoc Melih Sener, to piece together all the experimental evidence over the past decades and actually model a full chromatophore, down to every last atom. This work was made possible in huge part by Schulten's collaborator in Sheffield, Neil Hunter. In the intervening years, Sener and Schulten finally were able to pin down a model of the chromatophore, a requisite step. It would be impossible to illustrate all the interlocking processes in the chromatophore until its full structure was realized.
In the chromatophore there are five key protein players. The first two (light harvesting complex I and light harvesting complex II) act as antennae, and the third protein, the reaction center, converts the harvested energy into electron transfer. That leaves two remaining protein types that are necessary for the final conversion of sunlight to ATP. Those two are called the bc1 complex and ATP synthase. After light hits the antennae of the chromatophore, it is delivered to the reaction center, which uses that energy to push an electron down a circuit. Then a special molecule (a quinone) gets loaded up with two electrons and two protons and once it is stacked with protons and electrons (and thereby called a quinol), this loaded molecule swims through the oily membrane to the bc1 complex. The bc1 complex then pumps the quinol's protons from the outside of the spherical chromatophore to the inside of it. With these protons, then the well-known ATP synthase uses the proton gradient to drive production of ATP. (For completeness, there is a sixth protein involved, cytochrome, which shuttles electrons from bc1 back to the reaction center.)
Schulten's group had to utilize the tools of quantum chemistry to describe the complicated electron and proton transfer reactions that happen in the bc1 complex, efforts led most recently by graduate student Angela Barragan. She conducted very careful, large-scale quantum chemistry calculations to pin down what is actually happening in the process of creating a proton gradient.
As expected, the work Schulten's group undertook on ATP synthase is dear to his heart, as he puts it. It has taken him many years to achieve a suitable understanding of this membrane protein, ATP synthase. At first, it was slow going, for the initial structures were not complete enough to enable Schulten to dive fully into an explanation of its inner workings. “So then I was very careful and I published only the very little that I understood,” Schulten says about his work over the years on ATP synthase. “And today I finally think I have it nailed.” The reason Schulten believes he finally can explain ATP synthase and its function touches on the theme of his chromatophore movie as a whole: the teamwork of all the interlocking parts. ATP synthase has six proteins. Only today, with work most recently performed by a fantastic postdoc Abhi Singharoy, can Schulten describe the complexity of the system, namely how the six proteins work together to convert a proton gradient into a torque and then into actual ATP. While he still is verifying some of the steps used by ATP synthase, he is within reach of his childhood dreams about ATP. “Actually, I think I'm very close to what I would call my understanding the system.”
Photosynthesis Movie
The Photosynthetic Membrane of Purple Bacteria: A Clockwork of Proteins and Processes from TCBG on Vimeo.
The chromatophore movie thus encapsulates so many themes that have been winding through Schulten's brain over the past fifty years. He wanted to explore the molecules of life, especially the crucial ATP, ever since childhood. Thus he made the decision to study molecular physics in graduate school at Harvard, a discipline all his friends thought was boring and unworthy of Schulten's attention. In his first job he was exposed to the field of photosynthesis, but could only conduct limited studies on this topic, as the crucial membrane proteins at the heart of photosynthesis did not look like they would ever be solved. In Munich in the 1980s Schulten had a front row seat to solution of the first membrane protein, the reaction center, which turned his attention to the field of membrane protein simulation. But the reaction center had another profound effect, for it steered Schulten toward using the computer as a tool that, with honing, could become invaluable for biology. And finally, he has always pushed for simulating larger and larger structures with the explicit purpose of describing biological organization, which translates to looking at how teams of hundreds or even thousands of proteins interact. In fact, he had software tools he developed for computational biology, VMD and NAMD, and with those very tools he was capable of making the chromatophore movie, which required simulation and analysis of massive numbers of atoms (around 100 million). Keeping all this in mind, it is not surprising that Schulten claims, “the movie was also almost enforced on me.”
The chromatophore movie is a fitting closure to this history on membrane proteins, for it is a large, membrane-bound machine whose functionality was puzzled out to some degree with the aid of the computer. With the rich array of membrane proteins that Schulten and his group have demystified already, it is a wonder that Schulten had to wait almost thirty years until he could simulate a protein and membrane jointly. Admittedly, his road has not always been easy, but Schulten always looked forward and didn't dwell on the adversity. His successes in this field have gone hand in hand with the advances in computing tools that his group develops. In fact, with the use of the computer, Schulten has made a good living for himself even though getting structures of membrane proteins through crystallography is still somewhat slow going. While being a computational biologist is sometimes under appreciated, Schulten loves his life's calling. “So now I figure things out about living systems with my mathematical mind and I'm very, very happy with it.” For Schulten, then, pairing the membrane with a computer is the stuff dreams are made of.
Other Related Articles
If you enjoyed this article, you might be interested in these other histories:History of Magnetoreception
History of Polyenes
History of Bionanotechnology
History of Photosynthesis
History of NAMD
History of VMD
History of MDFF (Molecular Dynamics Flexible Fitting)
Comments?
For comments or inquiries, Lisa Pollack can be reached at the following: lpollack AT ks.uiuc.eduCitation
To cite this article, please use the following:Pollack, Lisa. "When Membrane Proteins Fuse with Computational Biology: A History" www.ks.uiuc.edu. 25 September 2014, from http://www.ks.uiuc.edu/History/membrane/
References
Hans-Joachim Werner, Klaus Schulten, and Albert Weller. Electron transfer and spin exchange contributing to the magnetic field dependence of the primary photochemical reaction of bacterial photosynthesis. Biochimica et Biophysica Acta, 502:255-268, 1978.
Oded Béjã et al., Bacterial Rhodopsin: Evidence for a New Type of Phototrophy in the Sea. Science 289:1902, 2000.
Dieter Oesterhelt and Walter Stoeckenius. Rhodopsin like protein from the purple membrane of Halobacterium halobium. Nature New Biology 233:149-152, 1971.
Dieter Oesterhelt and Walter Stoeckenius. Function of a new photoreceptor membrane. PNAS USA 70:2853-2857, 1973.
Walter Stoeckenius. The purple membrane of salt-loving bacteria. Scientific American. 234(6)38-46, 1976.
Richard Henderson. The Purple Membrane from Halobacterium Halobium. Ann. Rev. Biophys. Bioeng. 6:87-109, 1977.
"Hartmut Michel - Biographical". Nobelprize.org. Nobel Media AB 2014. Web. 29 Jul 2014. http://www.nobelprize.org/nobel_prizes/chemistry/laureates /1988/michel-bio.html
Helmut Heller, Michael Schaefer, and Klaus Schulten. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal-phases. Journal of Physical Chemistry, 97:8343-8360, 1993.
Gary A Taubes. The Rise and Fall of Thinking Machines. Inc., 15 Sep 1995.
Emad Tajkhorshid, Peter Nollert, Morten Ø. Jensen, Larry J. W. Miercke, Joseph O'Connell, Robert M. Stroud, and Klaus Schulten. Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science, 296:525-530, 2002.
Justin Gullingsrud, Dorina Kosztin, and Klaus Schulten. Structural determinants of MscL gating studied by molecular dynamics simulations. Biophysical Journal, 80:2074-2081, 2001.
Marcos Sotomayor and Klaus Schulten. Molecular dynamics study of gating in the mechanosensitive channel of small conductance MscS. Biophysical Journal, 87:3050-3065, 2004.
Marcos Sotomayor, Trudy A. van der Straaten, Umberto Ravaioli, and Klaus Schulten. Electrostatic properties of the mechanosensitive channel of small conductance MscS. Biophysical Journal, 90:3496-3510, 2006.
Valeria Vásquez, Marcos Sotomayor, Julio Cordero-Morales, Klaus Schulten, and Eduardo Perozo. A structural mechanism for MscS gating in lipid bilayers. Science, 321:1210-1214, 2008.
Wenjian Wang, Susan S. Black, Michelle D. Edwards, Samantha Miller, Emma L. Morrison, Wendy Bartlett, Changjiang Dong, James H. Naismith, Ian R. Booth. The Structure of an Open Form of an E. coli Mechanosensitive Channel at 3.45 Å Resolution. Science, 32:1179-1183, 2008.
James Gumbart and Klaus Schulten. Molecular dynamics studies of the archaeal translocon. Biophysical Journal, 90:2356-2367, 2006.
James Gumbart, Leonardo G. Trabuco, Eduard Schreiner, Elizabeth Villa, and Klaus Schulten. Regulation of the protein-conducting channel by a bound ribosome. Structure, 17:1453-1464, 2009.
Jens Frauenfeld, James Gumbart, Eli O. van der Sluis, Soledad Funes, Marco Gartmann, Birgitta Beatrix, Thorsten Mielke, Otto Berninghausen, Thomas Becker, Klaus Schulten, and Roland Beckmann. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nature Structural & Molecular Biology, 18:614-621, 2011.
Stephan Wickles, Abhishek Singharoy, Jessica Andreani, Stefan Seemayer, Lukas Bischoff, Otto Berninghausen, Johannes Soeding, Klaus Schulten, Eli van der Sluis, and Roland Beckmann. A structural model of the active ribosome-bound membrane protein insertase YidC. eLife, 3:e03035, 2014.
Anton Arkhipov, Ying Yin, and Klaus Schulten. Four-scale description of membrane sculpting by BAR domains. Biophysical Journal, 95:2806-2821, 2008.
Ying Yin, Anton Arkhipov, and Klaus Schulten. Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure, 17:882-892, 2009.
Hang Yu and Klaus Schulten. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS Computational Biology, 9:e1002892, 2013.
Fatemeh Khalili-Araghi, Emad Tajkhorshid, and Klaus Schulten. Dynamics of K+ ion conduction through Kv1.2. Biophysical Journal, 91:L72-L74, 2006.
Fatemeh Khalili-Araghi, Vishwanath Jogini, Vladimir Yarov-Yarovoy, Emad Tajkhorshid, Benoit Roux, and Klaus Schulten. Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophysical Journal, 98:2189-2198, 2010.



