Intrinsic Curvature Properties of Photosynthetic Proteins in Chromatophores
Protein-induced shaping of cellular membranes has enjoyed a great deal of interest in recent years. Common mechanisms of membrane shaping by proteins involve either protein scaffolds, e.g., BAR domains, or the shape and oligomerization of integral membrane proteins. An elegant example of membrane reshaping by membrane proteins is seen in the chromatophores of purple photosynthetic bacteria. Chromatophores are the bacterial photosynthetic pseudo-organelles which form as extensions of the cytoplasmic membrane upon transition to phototrophic growth and house the photosynthetic proteins. The shape of these chromatophores varies among species, the two most common forms being stacks of flat lamellar folds seen in Rhodopseudomonas (Rps.) acidophila and Rhodospirillum (Rs.) molischianum and small spherical vesicles in Rhodobacter (Rba.) sphaeroides and Rb. capsulatus (see Fig. 1). It is thought that the shape of the chromatophore is determined by the proteins it contains, light harvesting complexes 1 and 2 (LH1 and LH2), cytochrome bc1 (bc1), and possibly ATP synthase, and their interactions with the membrane and with each other.
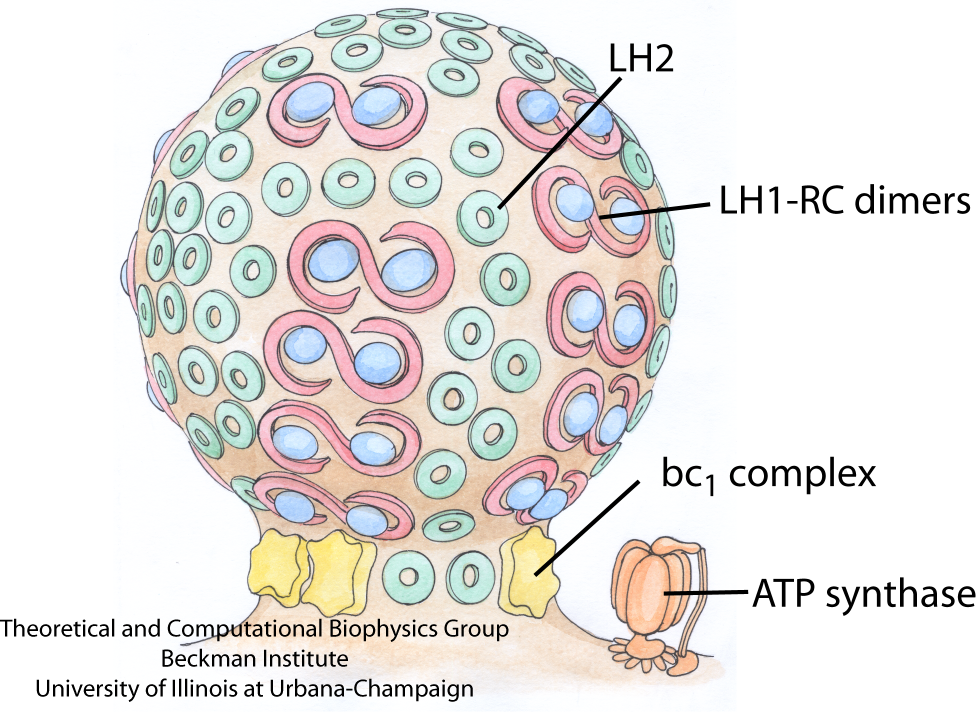
Fig. 1 - Schematic of the vesicular photosynthetic chromatophore. Shown here is a drawing of a chromatophore from Rba. sphaeroides. The LH1-RC dimers and LH2 complexes are closely packed in the bulb of the chromatophore, as seen in AFM images, and the bc1 complex and ATP synthase, which are absent from AFM images, are tentatively placed near the neck of the chromatophore.
Picture drawn by Olga Svinarski.
In a publication in 2008, we present the first set of simulations on the three chromatophore proteins, LH2, LH1-RC-PufX dimer, and bc1, designed to study their curvature properties at the atomistic resolution. Systems of seven LH2s pack and induce curvature in a way which depends on their packing density. A single LH1-RC-PufX dimer spontaneously bends at its dimerizing interface in a way which also induces curvature. bc1 does not induce curvature in simulations, neither singly nor in pairs. These simulations are the beginning of a study of much broader scope, which seeks to explore the interactions between many light harvesting proteins that lead to the creation of a chromatophore.
Recently, the availability of a new three-dimensional density map of the LH1-RC-PufX dimer made it possible to further refine the model of the complex. Using the Molecular Dynamics Flexible Fitting method developed in the group, two aspects of the LH1-RC-PufX dimer were elucidated in two 2009 articles: the light-absorbing and subsequent energy transferring features, and the membrane-bending properties.
Light-Harvesting Complex 2 (LH2)
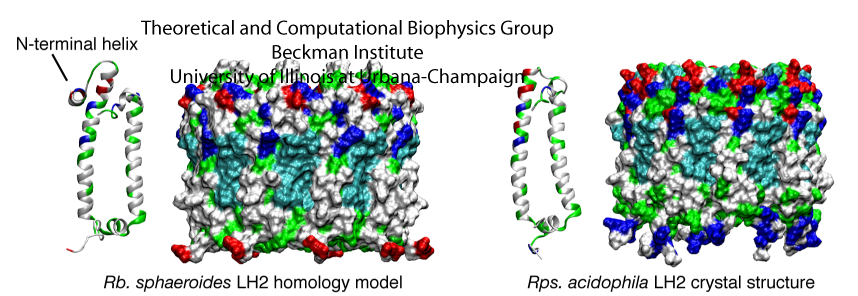
Mutant studies have implicated LH2 in producing the spherical shape of Rba. sphaeroides chromatophores. Further studies found that chromatophore size was inversely proportional to LH2 concentration, suggesting that more LH2 leads to stronger curvature. We sought to investigate via MD simulation whether LH2s from a species with vesicular chromatophores (Rba. sphaeroides) would have curvature properties different from LH2s from a species with lamellar chromatophores (Rps. acidophila). See Fig. 2 for a structural comparison between the LH2s of Rba. sphaeroides and Rps. acidophila.
Fig. 2 - Structural comparison of the Rps. acidophila LH2 crystal structure and the Rba. sphaeroides LH2 model. The proteins are shown in a surface representation, colored according to residue type (blue is basic, red is acidic, green is polar, and white is nonpolar). Figure made with VMD.
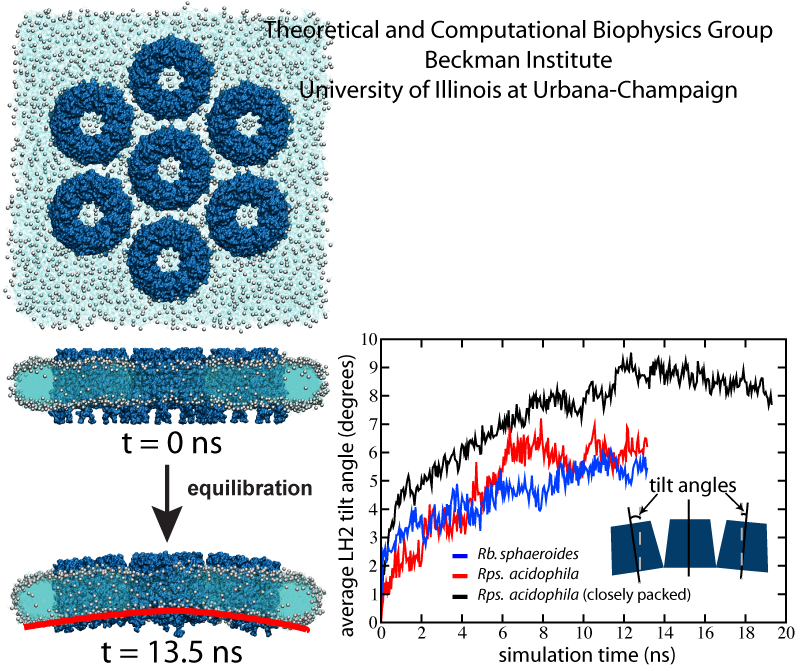
Seven LH2s were placed in the membrane in a hexagonally-packed arrangement, allowing just enough space between LH2s for a single line of lipids (Fig. 3). The simulation was performed once with Rps. acidophila LH2s and once with Rba. sphaeroides LH2s constructed via homology modeling to determine whether LH2 curvature properties would differ between the two species, possibly due to the small structural differences between the two LH2 models as shown in Fig. 2.
In both simulations, the LH2s packed together and tilted with respect to adjacent LH2s, producing a curved patch within a few nanoseconds, and stabilizing to a final curvature after approximately 10 ns (Fig. 3). Additionally, the lipids in contact with the LH2s adopted the same curvature as the LH2s, and the membrane as a whole adopted an overall curvature. No substantial differences were observed in the curvature of Rps. acidophila versus Rba. sphaeroides LH2s. The tilting angle of 6 degrees corresponds to approximately a radius of curvature of 790 Angstroms. A typical radius for a spherical chromatophore, by comparison, is approximately 150-400 Angstroms, smaller than that measured here. However, it is expected that the inclusion of more LH2s would lead to greater curvature.
That both species adopted approximately the same curvature was unexpected, as the chromatophores of Rps. acidophila are lamellar and not highly curved. The results suggest that the curvature properties of LH2 arise not from the structural differences, but rather from some other property that is conserved in both species. This question is currently under investigation.
The results of these simulations imply that LH2 complexes can induce curvature via packing, and that this behavior does not depend strongly on the small sequence or structural differences between LH2s from different species. However, LH2-induced curvature is amplified by closer packing of the proteins.
Fig. 3 - LH2-induced curvature. (Left) Equilibration simulation setup of the Rps. acidophila system with loose packing at t = 0 ns and at t = 13.5 ns. The LH2 complexes are shown in dark blue, with lipid head groups shown as grey spheres, and lipid tails in light blue. By the end of the 13.5 ns simulation, the LH2s tilt away from their neighbors to form a curved array, followed by lipid curvature. (Right) Plot of average tilt angle vs. time for the three simulations. The more sparsely packed Rps. acidophila and Rba. sphaeroides systems converge to final tilt angles of approximately 6 degrees. The closely packed Rps. acidophila system converged to a final tilt angle of approximately 8.5 degrees. Figure made with VMD.
LH1-RC-PufX (Core complex) Dimer -- Part I
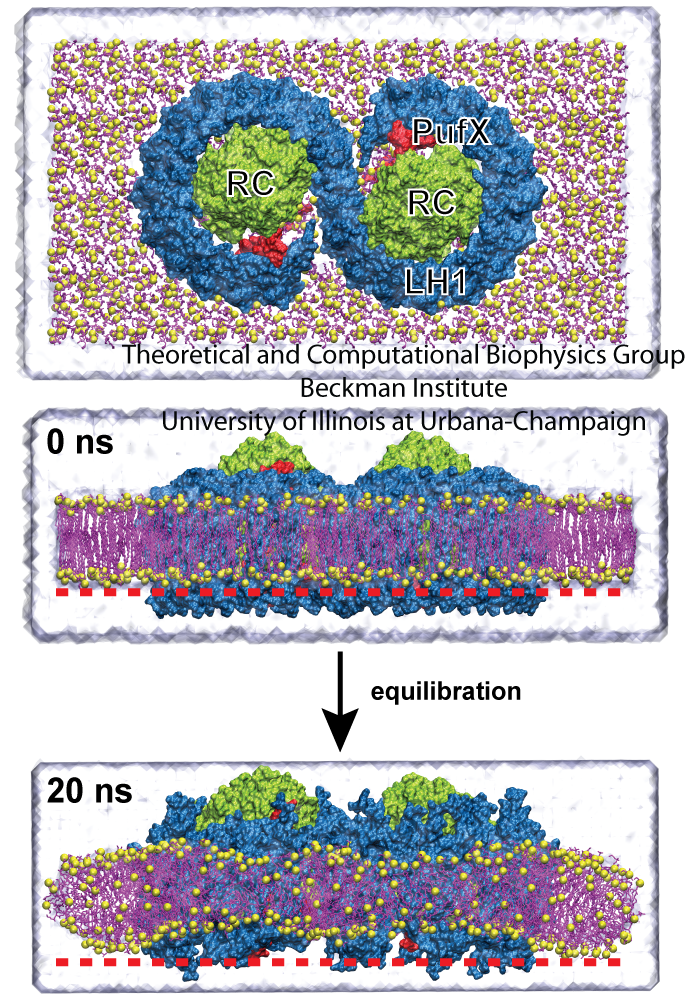
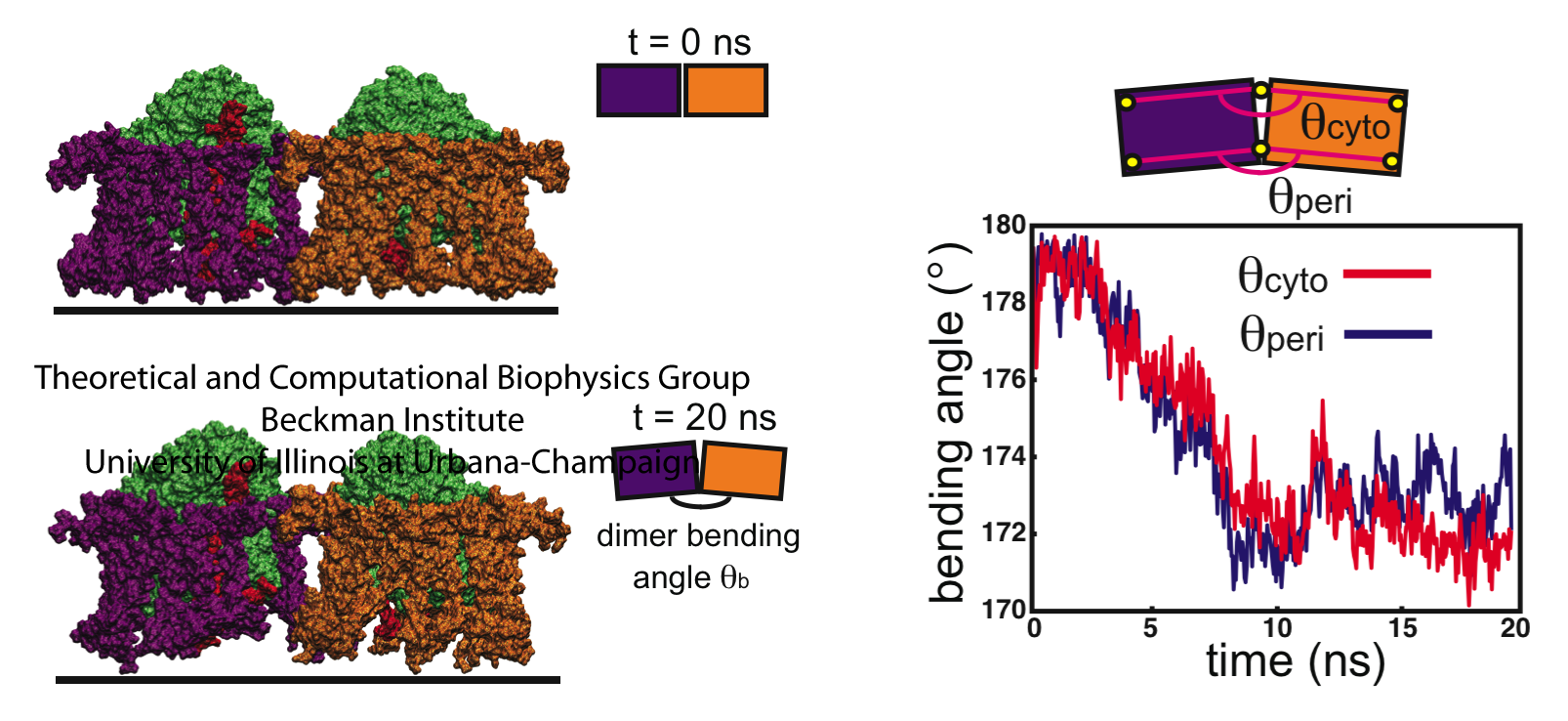
Mutant studies have shown that in LH2-deficient Rba. sphaeroides, tubular chromatophores form instead of spherical chromatophores, suggesting that the LH1-RC-PufX dimers generate cylindrical curvature. To test this hypothesized membrane-bending property of the Rba. sphaeroides LH1-RC-PufX dimer, a model dimer was constructed and equilibrated for 20 ns (see Fig. 4). Initially the dimer was flat when viewed along the membrane plane (due to the lack of 3D structural information at the time), but during the simulation, a bend at the dimerization junction was seen to develop (Fig. 4, bottom).
Fig. 4 - Simulation of the LH1-RC-PufX dimer. LH1 is colored in blue, the RCs in green, and PufX in red. The LH1 protein exhibits a slight bending after the 20 ns simulation. The membrane adopts to the LH1's change in shape. Figure made with VMD.
The observed LH1-RC-PufX bending angle is shown in Fig. 5: the bending angle developed quickly as simulation started and fluctuated around 172 degrees for the rest of the equilibration. The bending angle of 172 degrees corresponds to a radius of curvature of approximately 720 Angstroms. It is worth noting that the radius of the tubular chromatophores has been estimated to be 250-550 Angstroms, which corresponds to a dimer bending angle of 160-170 degrees. Therefore, the bending angle of LH1-RC-PufX measured here seems to be insufficient to induce the correct membrane curvature.
Fig. 5 - Bending of the LH1-RC-PufX dimer. The initially "flat" LH1-RC-PufX dimer exhibits bending at the dimerizing junction after equilibration. Figure made with VMD.
LH1-RC-PufX (Core complex) Dimer -- Part II
As discussed previously, in case of the RC-LH1-PufX dimer, the bending seen in the simulations was less than what was measured in tubular vesicles, i.e., a full explanation of RC-LH1-PufX-induced membrane curvature seemed to be lacking. Recent electron microscopy (EM) single-particle analysis of the negatively stained Rba. sphaeroides RC-LH1-PufX dimer at 25 Angstroms resolution showed, though, that the RC-LH1-PufX dimer is more prominently bent than seen in the simulations, with a bending angle of 146 degrees. We therefore sought to combine the structural information provided in the EM map with the all-atom model developed in Chandler et al., employing for this purpose the methodology described in Trabuco et al.. This methodology, termed Molecular Dynamics Flexible Fitting (MDFF), accelerates dimer bending, and thereby permits one to observe the effect of the bent dimer on membrane geometry.
The result of the MDFF simulations is best illustrated through the trajectory: click here for the simulation trajectory (27M). From the simulation, it was observed that the RC-LH1-PufX dimer was quickly fitted into the transparent EM density, and the whole protein became bent. After 5 ns of MDFF, the system was allowed to equilibrate freely, and during the equilibration the membrane curvature continued to develope. At the end of the simulation, the membrane became an arched patch, and one also recognizes that there is a little "twist" in the membrane besides the bending.
To see the developement of the membrane geometry better, for each frame of the above trajectory a best-fit surface was generated for the membrane patch, resulting in a movie showing how the membrane curvature formed: click here for the movie (17M). One can also compute the radius of curvature at each point on the membrane surface, and color the surface accordingly. By doing so, the geometry of the membrane can be quantified, and it can be recognized that the axis of maximum curvature (colored red in the movie) is slanted in respect to the dimer short axis, indicating the asymmetry of the membrane patch.
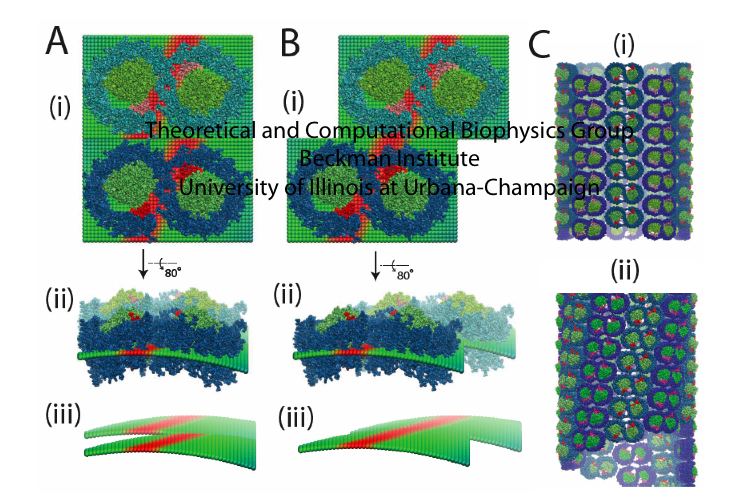
The local curvature properties seen in simulation actually is related to the long-range arrangement of the protein. The packing arrangement of RC-LH1-PufX was explored by constructing two all-atom tubular vesicles, one built with the RC-LH1-PufX dimers stacked along their short dimer axes, and the other built by stacking the dimers with a slight offset. Compared to the EM image of a native RC-LH1-PufX tube from LH2-deficient Rba. sphaeroides, which shows that the dimers are organized in a helical arrangement, the tubular vesicle matches the EM image better. Therefore, the "offset" stacking arrangement of RC-LH1-PufX dimers that leads to the helical organization of the dimers, i.e., a global effect, can be explained by the twisting of the RC-LH1-PufX dimer's local curvature. These results are presented in more detail in Hsin et al., 2009.
Fig. 6 - Local curvature properties of the RC-LH1-PufX dimer and the long-range organization
of native Rba. sphaeroides tubes. (A) In a stacking arrangement in which LH1-RC-PufX dimers
are placed directly on top of each other, their local curvatures cannot complement each other.
(B) In a stacking arrangement in which dimers are placed with a slight offset that aligns their
maximum curvature axes, their local curvature effects are complementary and form a cylindrical
surface. (C) Two LH1-RC-PufX tubular vesicles were constructed, one with the "lined-up" stacking
arrangement, and one with the "offset" stacking arrangement. The second arrangement (C-ii)
better resembles the recent EM image of the native RC-LH1-PufX tubular vesicle. Figure made with VMD.
Cytochrome bc1 (bc1)
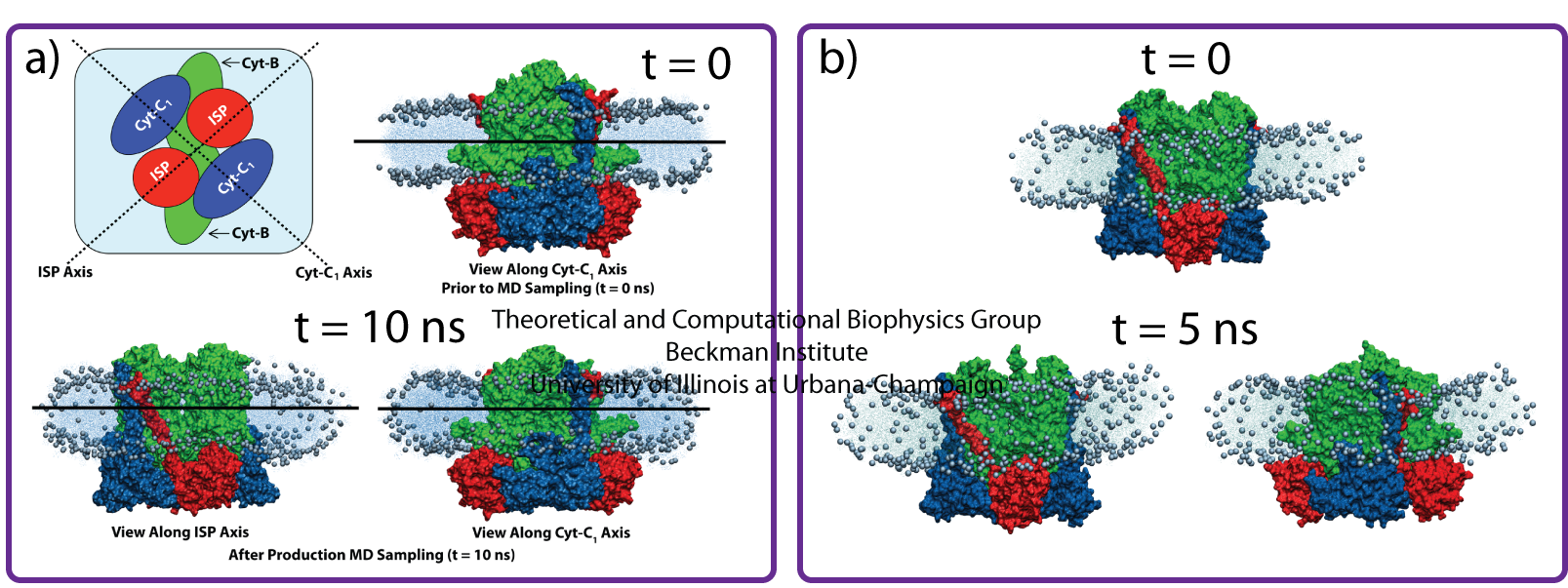
AFM and electron microscopy images reveal location and inter-protein organization of the LH2 and LH1-RC-PufX complexes in Rba. sphaeroides chromatophores, but did not identify the locations of the bc1 complex. The determination of the lipid bilayer curvature surrounding a functional dimer of bc1 complexes offers potential insight into the location of bc1 within the photosynthetic chromatophores. Simulations of single and multiple bc1 dimers were conducted to examine the first of these two scenarios. The following results are presented in more detail in Chandler et al., 2008.
A single bc1 dimer embedded in a membrane patch was solvated and equilibrated for 10 ns (Fig. 9a). After the simulation, as seen in Fig. 9a, the lipid bilayer surrounding the single bc1 dimer remained flat. When this simulation was repeated, the bc1 protein/membrane system result in a negatively curved lipid (Fig. 9b). This lipid curvature was observed to occur subsequent to a specific structural change within the bc1 complex early in the simulation, and shows that the bc1 dimer can occupy a curved as well as flat lipid bilayer.
Fig. 9 - Results from simulations of a single bc1 dimer from Rba. sphaeroides, showing that the bc1 dimer can occupy a curved as well as flat lipid bilayer. Figure made with VMD.
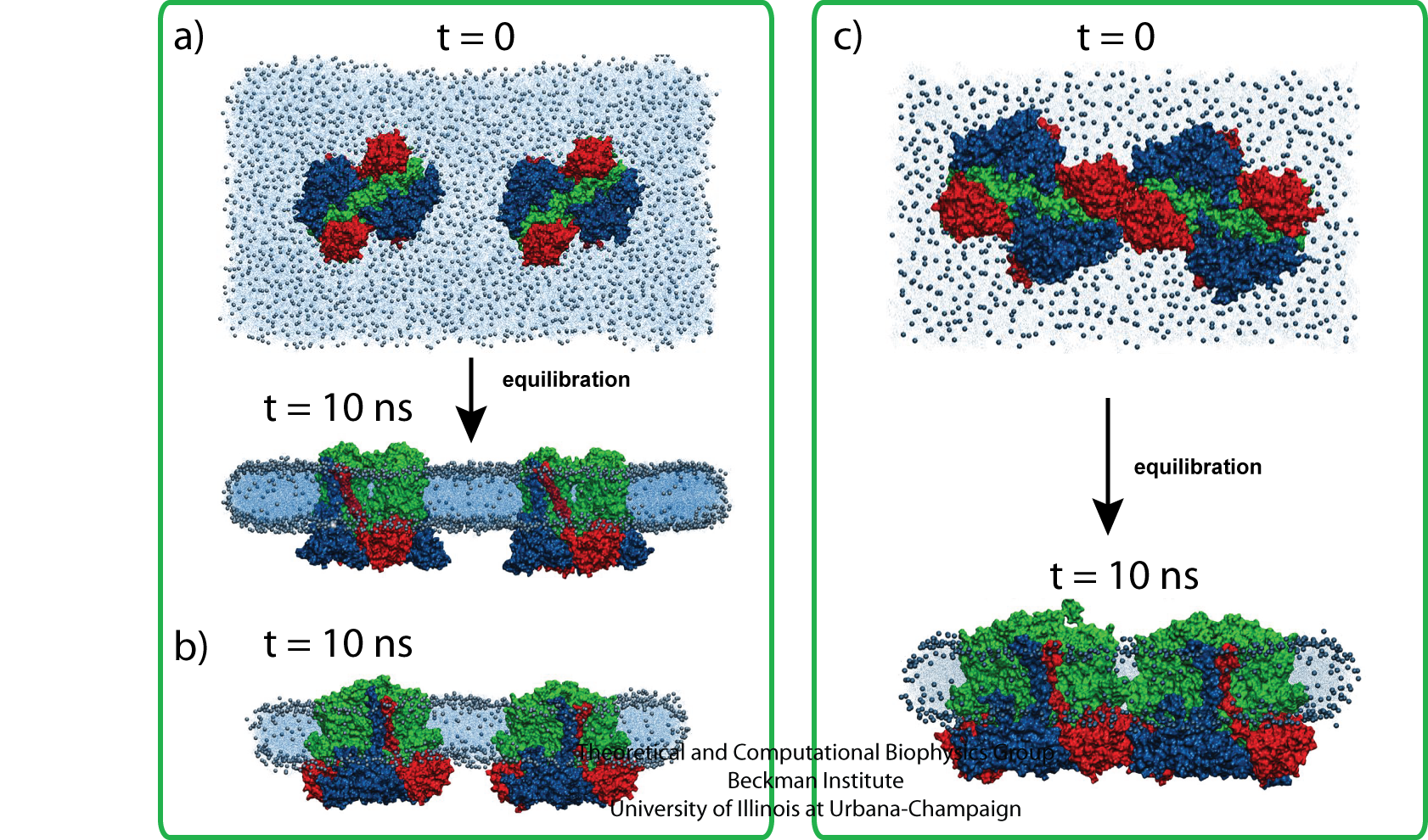
A second set of simulations were conducted to investigate the possibility that multiple bc1 dimers cooperatively promote curvature formation in the membrane region separating the proteins. Three simulations with two bc1 dimers were performed with different bc1 arrangements. No curvature was observed in all three simulations, shown in Fig. 10. Taken together, these equilibration simulations suggest that although bc1 dimers may accommodate curved membranes, they do not themselves directly induce membrane curvature through bc1-bc1 or bc1-membrane interactions.
Fig. 10 - Simulations with two bc1 dimers with three different bc1 arrangements; no curvature was seen. Figure made with VMD.
Also see a related press article "The Computational Microscope. Ranger's parallel computing power enables researchers to model the largest biomolecular apparatus to date" at TACC - Texas Advanced Computing Center, or HPCWire
.
Related publications
Membrane curvature induced by aggregates of LH2s and monomeric LH1s.Danielle E. Chandler, James Gumbart, John D. Stack, Christophe Chipot, and Klaus Schulten. Biophysical Journal, in press (2009).
Protein-induced membrane curvature investigated through molecular dynamics flexible fitting.
Jen Hsin, James Gumbart, Leonardo G. Trabuco, Elizabeth Villa, Pu Qian, C. Neil Hunter, and Klaus Schulten.
Biophysical Journal, 97:321-329 (2009).
Intrinsic Curvature Properties of Photosynthetic Proteins in Chromatophores.
Danielle Chandler, Jen Hsin, Christophore B. Harrison, James Gumbart, and Klaus Schulten.
Biophysical Journal, 95:2822-2836 (2008).
Investigators
Collaborators
Pu Qian (University of Sheffield)
Neil Hunter (University of Sheffield)
John D. Stack (UIUC)
Related web pages on our site
Chromatophore and photosynthesis:
Structure and function of the photosynthetic core complex
Atomic level structure and function of a photosynthetic vesicle
Organization of energy transfer networks.
Quantum Biology of the Photosynthetic Unit
Light-Harvesting by Carotenoids
Protein-induced membrane curvature:
Membrane Sculpting by BAR domains
Page created and maintained by Jen Hsin.