Neurons and Synapses Spotlights
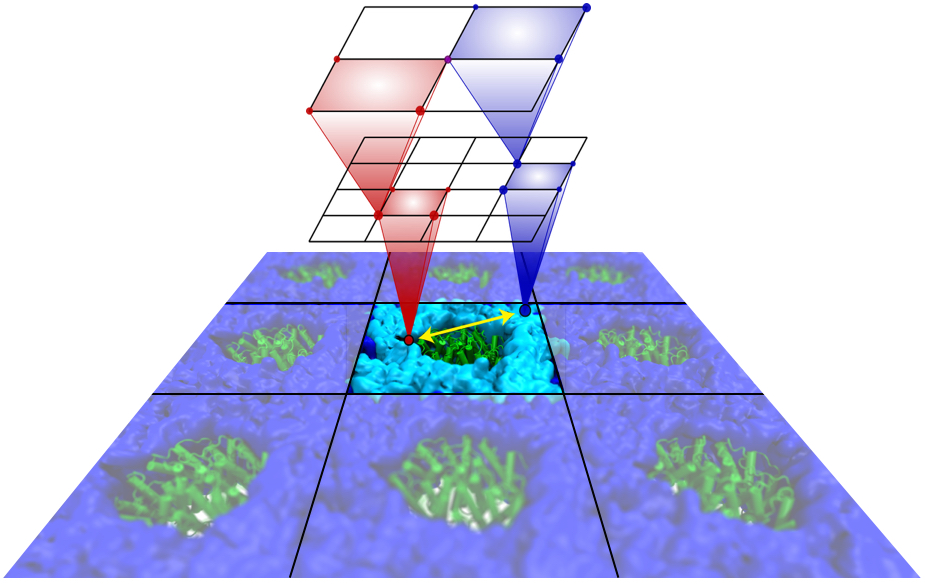
Long-range electrostatic interactions control macromolecular processes within living cells as prominent charges appear everywhere, such as in DNA or RNA, in membrane lipid head groups, and in ion channels. Reliable and efficient description of electrostatic interactions is crucial in molecular dynamics simulations of such processes. Recently a new mathematical approach for calculating electrostatic interactions, known as multilevel summation method (MSM), has been developed and programmed into NAMD 2.10 as reported here. Compared to the earlier decades-long approach, the particle-mesh Ewald (PME) method, MSM provides more flexibility as it permits non-periodic simulations like ones with asymmetric charge distributions across a membrane or of a water droplet with a protein folding inside. Furthermore, MSM is ideally suited for modern parallel computers, running, for example, simulations of large virus particles. More information here.
image size:
258.8KB
made with VMD
Neurons in the brain form a closely knit communication network with other neurons. Each neuron sends messages through up to thousands of cell-cell communication channels, so-called synapses. To avoid communication chaos, the messages of each neuron are chemically encoded as if neurons speak English to some neurons and French to others. The neurons employ an extremely efficient encoding system, packaging chemical message molecules, so-called neurotransmitters, in spherical vesicles encapsulated by a lipid membrane just like the whole neuron is encapsulated by a membrane. The vesicles aggregate near the presynaptic site of the membrane, ready to release their neurotransmitters into the space between neurons at the synapse, the so-called synaptic cleft. When a sender neuron becomes electrically active, as it wants to "speak", the electrical activity releases Calcium ions at the pre-synaptic cell that trigger merging (fusion) of vesicles with the sender neuron's membrane. At this point the neurotransmitter molecules flow into the synaptic cleft. The receiver neuron "hears" the signal by receiving the neurotransmitter molecules on receptors in the postsynaptic membrane, inducing as a result an electrical signal in the receiver neuron. The release involves a group of proteins that make vesicles ready for the release and proteins that execute the Calcium-triggered step, among the latter synaptotagmin I. As reported recently, researchers have proposed with the help of computer simulations using NAMD how synaptotagmin I acts. The simulations suggest that the experimentally observed structure of synaptotagmin I measured in vitro in a crystal/NMR form of the protein differs from the active, in situ form of synaptotagmin I. The finding, if true, will be a dramatic example for the role of computing in biology where the computer often complements observation studying, as in the present case, biomolecules in situ, namely their natural environment, rather than in vitro, namely in an artificial environment. Please read more on our neuron transmission website.
image size:
151.9KB
made with VMD
For many, the word 'X-ray' conjures up the images of white bones on black backgrounds hanging on the wall of a doctor's office. However, X-rays have played another important role for the past 100 years through their use in the determination of chemical structures at atomic level detail, starting with the first ever structure of table salt in 1924. Since then, the diffraction properties of X-rays, when shone on a crystal, have been used to solve increasingly large and complex structures including those of biological macromolecules found inside living cells. X-ray crystallography has become the most versatile and dominant technique for determining atomic structures of biomolecules, but despite its strengths, X-ray crystallography struggles in the case of large or flexible structures as well as in the case of membrane proteins, either of which diffract only at low resolutions. Because solving structures from low-resolution data is a difficult, time-consuming process, such data sets are often discarded. To face the challenges posed by low-resolution, new methods, such as xMDFF (Molecular Dynamics Flexible Fitting for X-ray Crystallography) described here, are being developed. xMDFF extends the popular MDFF software originally created for determining atomic-resolution structures from cryo-electron microscopy density maps (see the previous highlights Seeing Molecular Machines in Action, Open Sesame, Placing New Proteins, and Elusive HIV-1 Capsid). xMDFF provides a relatively easy solution to the difficult process of refining structures from low-resolution data. The method has been successfully applied to experimental data as described in a recent article where xMDFF refinement is explained in detail and its use is demonstrated. Together with electrophysiology experiments, xMDFF was also used to validate the first all-atom structure of the voltage sensing protein Ci-VSP, as also recently reported. More on our MDFF website.
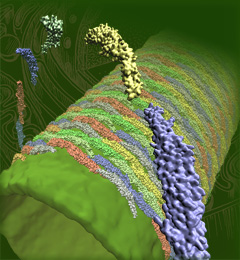
image size:
1.3MB
Olga Svinarski and VMD
The cells of higher life forms, so-called eukaryotic cells, are subdivided through many internal membranes made of lipid bilayers. The internal membranes assume numerous shapes, like spheres, tubes or parallel sheets. Outside of cells, biological membranes adopt usually flat shapes and the question arises, how do eukaryotic cells sculpt their inner membranes? The question of flat membrane sculpting is particularly interesting also as mature cells constantly produce new membrane shapes, for example spherical vesicles filled with certain biomolecules destined for release into the extracellular space, a process called exocytosis. The cell has many mechanisms available for sculpting its membranes, one of them relying on proteins called BAR domains that act from the surface of lipid bilayers. Molecular modeling with NAMD and VMD has provided valuable views of BAR domains at work in case of the so-called N-BAR family (see the earlier highlights Protein Teamwork, Jun 2009 and Proteins Sculpting Cell Interior, Sep 2008). Researchers report now an extension of the earlier studies to the F-BAR domain family of membrane sculpting proteins. The new modeling work is particularly exciting as it can be directly compared to electron microscopy images of membrane tubes sculpted from flat membranes in experiments done outside of cells. The new studies reveal how F-BAR domains sculpt tubular membranes through the shape of dimerized domains and through F-BAR domains not acting individually, but as an army of F-BAR domains adopting an ordered formation on one side of the membrane. More on our F-BAR domain web page.
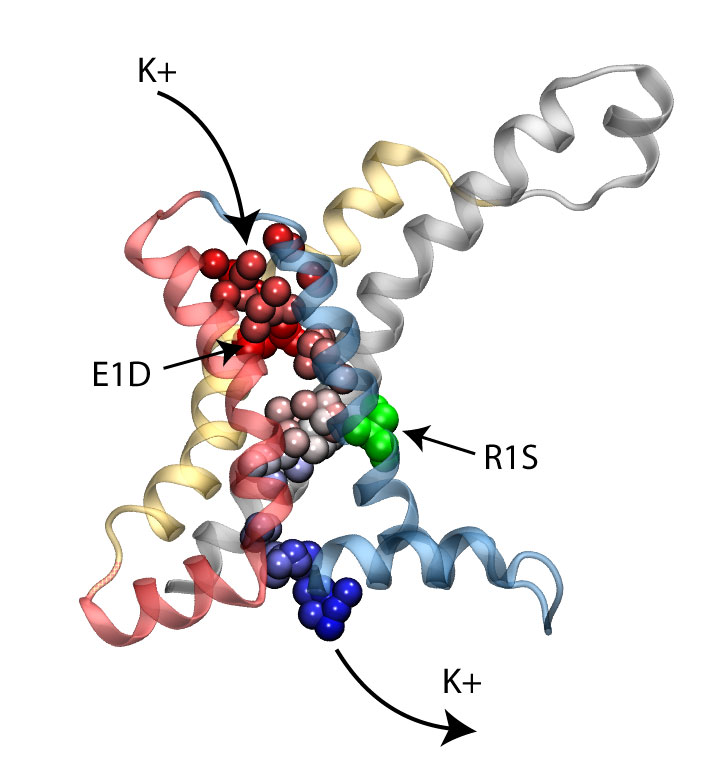
image size:
419.0KB
made with VMD
Voltage-gated ion channels, present in the membrane of excitable cells, control the ionic concentrations of the cellular environment by maintaining a potential difference of -100 mV between inside and outside of the cell membrane. Voltage-sensing occurs through distinct protein modules, known as voltage-sensor domains, four of which surround the main conduction pathway in potassium channels. Mutation of a certain amino acid on the voltage sensor domain turns these protein modules into cation channels, known as omega pores, which allow conduction of ions only when the main pathway is closed. Omega pores closely resemble the long-sought voltage-gated proton channels, which were recently identified to follow the same voltage-sensing mechanism as voltage-gated cation channels. In a recent report, researchers have visualized the twisted permeation pathway of the ions through omega pores using the molecular dynamics program NAMD. The simulations revealed a narrow constriction region lined by negatively charged amino acids, acting as a selectivity filter that prefers passage of positively charged ions through the pore. For more detail, see our potassium channel website .
Nerve cells, through their electrical signals, control actions and intelligence of higher organisms. The signals result mainly from potassium and sodium ion channels in the cells: when the cells are stimulated electrically, they send an all (in case of sufficient stimulation) or nothing signal to other nerve cells or organs like muscle. As shown in ground breaking work by 1963 Nobelists Hodgkin and Huxley, cast into mathematical equations, nerve cells establish these signals through voltage gating of channels. The nature of the gating, monitored through the so-called gating current, has been elusive for decades, despite a detailed characterization of the ion conductivity itself rewarded through a 2003 Nobel Prize to MacKinnon. The riddle is that the channel involves a protein with few charged amino acids that seem to be only weakly coupled energetically to an electrical potential gradient across the cell membrane. Now a sweeping modeling study using NAMD employing the most powerful computers available to researchers today has led to an explanation of voltage-gating. Simulations revealed that the potential gradient is focused by the channel protein to a very narrow region such that its value is much larger than anticipated. The protein was also seen to arrange its charged amino acids sensing the gradient in an unusual helix, a so-called 310 helix, that aligns charges perfectly while at the same time inducing a motion that opens and closes the channel. Proof of the veracity of the computational model is that the calculated gating current fits perfectly the observation. More on our potassium channel website.

image size:
287.1KB
made with VMD
Living cells organize many functions: molecular import and export, signaling, transcription of genes into proteins, movement, building, repair, and more. These functions are realized through a complex architecture of the cell interior reflected in a system of labyrinthine membranes forming manifold cellular organelles from tubes, vesicles and many other shapes. Accordingly, cells need to sculpt their membrane in never ceasing processes and have at their disposal a wide range of mechanisms. A key sculpting mechanism is furnished by proteins, so-called BAR domains, that apparently form lattice-like scaffolds adhering to membrane surfaces. Such scaffolds have been observed through electron microscopy and they are now also being described through molecular dynamics simulations using NAMD. As recently reported, the simulations, carried out at four different levels of resolution (from an atomic to a continuum level), revealed that different arrangements of BAR domains lead to different curvatures. The simulations help to explain why BAR domains working in teams, i.e., in lattice formation, sculpt intra-cellular membranes into different shapes, depending on the exact arrangement. An arrangement of BAR domains that is particularly efficient in bending membranes was identified. More information can be found on our BAR domain web page.
Biological cells, in particular neurons, maintain an inside-outside voltage gradient through active transport of ions (Na+, K+, Cl-, and others) across their membranes. The flow of the ions down their gradients through membrane channels is highly selective for each ion. The high selectivity permits nerve cells to signal each other through voltage spikes, which are produced through transient changes of channel conductivities for Na+ ions (channels open and close in about a ms) and K+ ions (channels open and close in about 10 ms). Crucial for the generation of voltage spikes is the selective, yet quick, conduction of ions, but as one knows from personal experience at border crossings, high selectivity and quick crossing seem to be mutually exclusive. Yet biological ion channels reconcile selectivity and speed. Prior experimental work, primarily that of year 2003 Nobelist MacKinnon, as well as computational work suggested how potassium channels achieve selectivity and speed. But until recently no high resolution atomic structure of a potassium channel was known in the open form and the suggested mechanism could not be tested under natural conditions through atomic level simulations. Last year's solution of the structure of the potassium channel Kv1.2 in its open form made it finally possible to simulate, using NAMD, the conduction of ions through Kv1.2 driven by a voltage gradient. The results reported recently confirmed indeed the high selectivity - high speed mechanism suggested earlier, namely a billiard-type motion of two and three ions, the last ion kicking the first ion out. The simulations revealed for the first time, through movies, the overall permeation process, including the jumps of ions between energetically favorable binding sites and the sequence of multi-ion configurations involved in permeation. More on our potassium channel web site.