Shashank Pant
I develop and use classical mechanical simulation methods to study the kinetics and thermodynamics of complex biological processes. I am specifically interested in performing all-atom simulations that can reach timescales of milliseconds and longer, with the aim to bridge gap between theory and experiments. As these timescales are far beyond the reach of the fastest supercomputers, my work entails the use of variety enhanced sampling algorithms to understand the dynamics of biomedically relevant biomolecules. Currently, I am working on characterizing the transition cycle of human glutamate transporters using advanced simulation techniques like: ConstantpH, Metadynamics, SMD, and Pathway sampling methods. Apart from this I also work actively on understanding the role of membrane environment and inhibitors in modulating the structure-function relationship of membrane proteins like: P-glycoproteins, Potassium-channels, and signalling domains.
Home Department: Biophysics and Quantitative Biology
Office Address:
Beckman Institute for Advanced Science and Technology, Room 3047
University of Illinois at Urbana-Champaign,
405 N. Mathews Ave,
Urbana, IL 61801,
Office Phone: +1 (217) 979-5572
Email Address: pant5@illinois.edu
Beckman Institute Graduate Fellow (2019)
Education
- BS-MS in Chemical Sciences , Indian Institute of Science Education and Research, Kolkata, India, 2015
Research Interests
- Large-Scale conformation changes in membrane transport proteins
- Specific lipid-protein interactions
- Developing statistical and machine learning based enhanced sampling methods
Publications ( Google Scholar )
- Glutamate transporters have a chloride channel with two hydrophobic gates, I. Chen*, S. Pant* , Q. Wu*, R. Cater, M. Sobti, R. Vandenberg, A. G. Stewart, E. Tajkhorshid, J. Font, and R. Ryan (2021), Nature, in press.
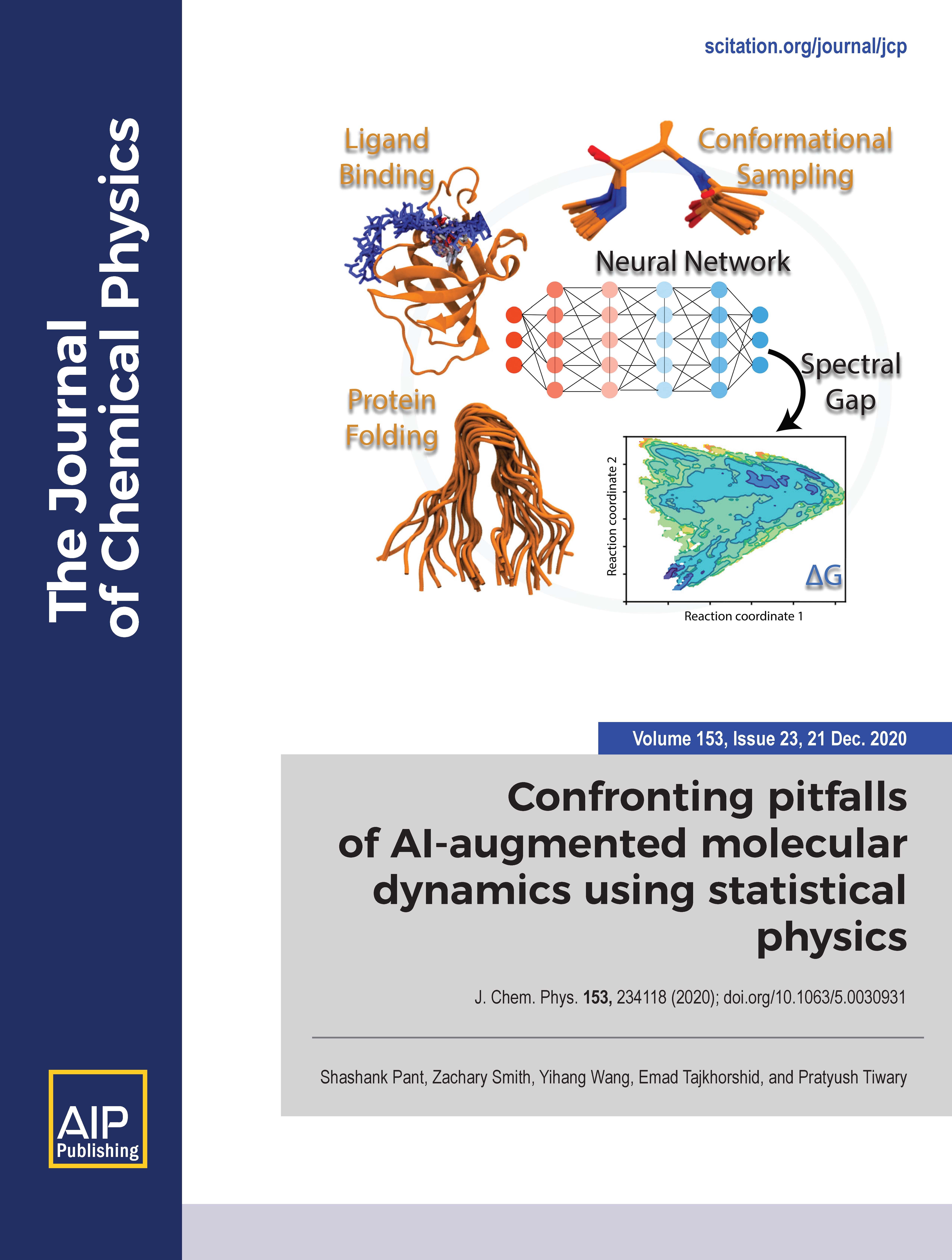
- Confronting pitfalls of AI-augmented molecular dynamics using statistical physics, S. Pant , Z. Smith, Y. Wang, E. Tajkhorshid, and P. Tiwary (2020), Journal of Chemical Physics, 154: 234118. (Cover Article) (Featured Article)
- Membrane Surface Recognition by the ASAP1 PH Domain and Consequences for Interactions with the small GTPase Arf1, O. Soubias*, S. Pant *, F. Heinrich, Y. Zhang, N. S. Roy, J. Li, X. Jian, M. E Yohe, P. A. Randazzo, M. Losche*, E. Tajkhorshid*, and R. A. Byrd* (2020), Science Advances, 6:eabd1882.
- Hydrogen-deuterium exchange mass spectrometry captures distinct dynamics upon substrate and inhibitor binding to a transporter, R. Jia, C. Martens, M. Shekhar, S. Pant , G. A. Pellowe, A. M. Lau, H. E. Findlay, N. J. Harrisa, E. Tajkhorshid, P. J. Booth, and A. Politis (2020), Nature Communications, in press.
- Microscopic Characterization of GRP1 PH Domain Interaction With Anionic Membranes, S. Pant and Emad Tajkhorshid (2020), Journal of Computational Chemistry, 41: 489-499. (Cover Article)
- Identifying mutation hotspots reveals pathogenetic mechanisms of KCNQ2 epileptic encephalopathy, J. Zhang, E. C. Kim, C. Chen, E. Procko, S. Pant , K. Lam, J. Patel, R. Choi, M. Hong, D. Joshi, E. Bolton, E. Tajkhorshid, and H.-J. Chung (2020), Scientific Reports, 10, 4756.
- Computational Dissection of Membrane Transport at a Microscopic Level, Tao Jiang, Po-Chao Wen, Noah Trebesch, Zhiyu Zhao, S. Pant , Karan Kapoor, Mrinal Shekhar, Emad Tajkhorshid (2020), Trends in Biochemical Sciences, 45: 202-216.
- Characterization of Lipid-Protein Interactions and Lipid-mediated Modulation of MembraneProtein Function Through Molecular Simulation, M. P. Muller*, T. Jiang*, C. Sun, M. Lihan, S. Pant , P. Mahinthichaichan, A. Trifan, and E. Tajkhorshid (2019), TOP5 community's Favorite Review of 2019
Chemical Reviews, 119: 6068-6161. - Confronting pitfalls of AI-augmented molecular dynamics using statistical physics, S. Pant , Z. Smith, Y. Wang, E. Tajkhorshid, and P. Tiwary (2020), Journal of Chemical Physics, 154: 234118. (Cover Article) (Featured Article)
TCBG
- Impact of length scale of attraction on the dynamical heterogeneity: a molecular dynamics simulation study,
S Pant , Pradip K Ghorai (2019),
Journal of Chemical Science, 131 (4) (COVER ARTICLE)
- Breakdown of 1D water wires inside charged carbon nanotubes
S Pant* (2016),
Chemical Physics Letters, 644, 159-166
- Structural anomaly of core-softened fluid confined in single walled carbon nanotube: a molecular dynamics simulation investigation
S Pant , PK Ghorai (2016),
Molecular Physics,114 (11), 1771-1777
- Effect of attractive interactions on the water-like anomalies of a core-softened model potential
S Pant , T Gera, N Choudhury (2013),
Journal of Chemical Physics,139, 244505
- Controlled synthesis of amino acid-based pH-responsive chiral polymers and self-assembly of their block copolymers
K Bauri, SG Roy, S Pant, P De (2013),
Langmuir, 29 (8), 2764-2774
- Dual pH and temperature responsive helical copolymer libraries with pendant chiral leucine moieties
K Bauri, S Pant, SG Roy, P De (2013),
Polymer Chemistry, 4 (14), 4052-4060
* corresponding author
Pre-TCBG
Cover Images