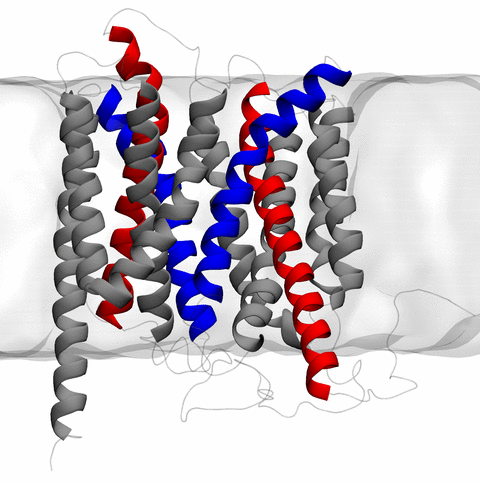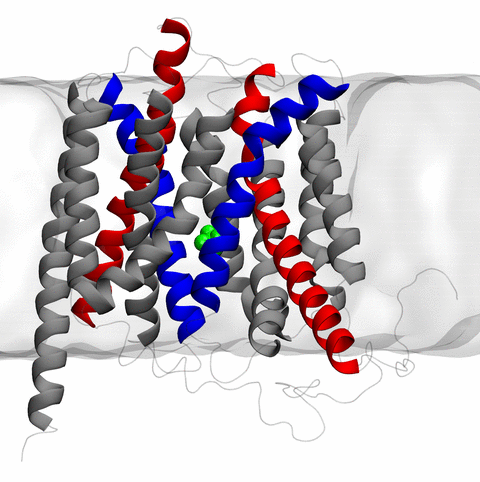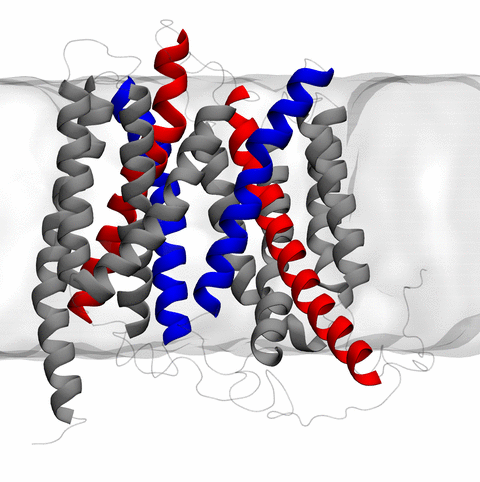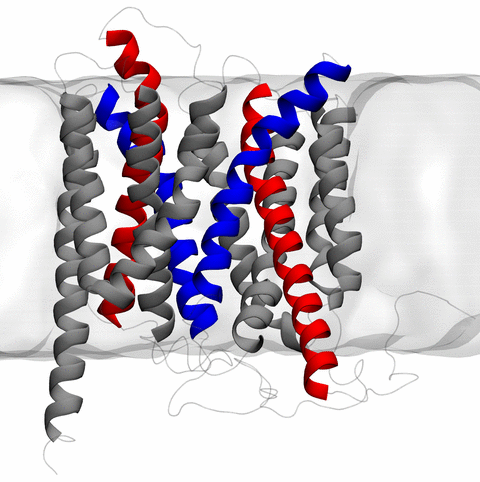
Home Codes and Scripts Movies and Videos Computational Structural Biology and Molecular Biophysics at UIUC

Sep 29, 2015 - My recent work was published in Nature Communications.
Aug 17, 2015 - I joined the faculty of the Department of Chemistry and Biochemistry at the University of Arkansas, Fayetteville .
Oct 10, 2014 - I have joined the Editorial Board for Scientific Reports (Nature Publishing Group).
Jan 28, 2014 - Transporter Protein Dance Moves: Biomedical Beat (Reasearch news from NIGMS, NIH) blogs about MsbA paper.
Dec 2, 2013 -
Difficult dance steps: Team learns how membrane transporter moves.

Mahmoud Moradi
Postdoctoral Associate
Computational Structural Biology and Molecular Biophysics
Department of Biochemistry and Beckman Institute
University of Illinois at Urbana-Champaign
Beckman #3015
Lync Phone: (217) 300-6722
Email Address: moradi@illinois.edu
Education
-
Ph.D., Physics, 2005-2011
NC State University, Raleigh, NC
Thesis: Free energies, transition mechanisms, and structural characteristics of proline-rich peptides. -
M.Sc., Physics, 2002-2004
Sharif University of Technology, Tehran, Iran
Thesis: On the probability of clusters in Abelian Sandpile Model. -
B.Sc., Physics, 1997-2001
Sharif University of Technology, Tehran, Iran
Research Interests
- Conformational transitions in membrane transport proteins
- ABC transporters, MFS transporters, EAATs
- Methodological developments for biomolecular simulations
- Enhanced sampling techniques, Path-finding algorithms
- Nonequilibrium statistical mechanics
- Nonequilibrium work relations, Adaptive-bias methods
Publications
2015
- J Li, P-C Wen, M Moradi, and E Tajkhorshid Computational Characterization of Structural Dynamics Underlying Function in Active Membrane Transporters. Curr. Opin. Struct. Biol., 31:96, 2015
- M Moradi, G Enkavi, and E Tajkhorshid, Atomic-level characterization of transport cycle thermodynamics in glycerol-3-phosphate transporter. Nat. Commun., 2015. (accepted)
2014
- M Moradi, C Sagui, and C Roland, Investigating rare events with nonequilibrium work measurements: I. nonequilibrium transition path probabilities. J. Chem. Phys., 140:034114, 2014.
- M Moradi, C Sagui, and C Roland, Investigating rare events with nonequilibrium work measurements: II. transition and reaction rates. J. Chem. Phys., 140:034115, 2014.
- R E Hulse, J Sachleben, P-C Wen, M Moradi, E Tajkhorshid, E Perozo, Conformational dynamics at the inner gate of KcsA during activation. Biochemistry (Rapid Report), 53:2557, 2014.
- M Moradi, and E Tajkhorshid, Computational recipe for efficient description of large-scale conformational changes in biomolecular systems. J. Chem. Theory Comput., 10:2866, 2014.
- G Enkavi, J Li, P-C Wen, S Thangapandian, M Moradi, T Jiang, W Han, and E Tajkhorshid, A microscopic view of the mechanisms of active transport across the cellular membrane. Annual Reviews in Computational Chemistry, 10:77, 2014.
2013
- M Moradi, and E Tajkhorshid,
Mechanistic picture for conformational transition of a membrane transporter at atomic resolution.
Proc. Natl. Aca. Sci. USA, 110:18916, 2013.



- M Moradi, and E Tajkhorshid, Driven metadynamics: Reconstructing equilibrium free energies from driven adaptive-bias simulations. J. Phys. Chem. Lett., 4:1882, 2013.
- M Moradi, V Babin, C Roland, and C Sagui, Reaction path ensemble of B-Z-DNA transition: A Comprehensive atomistic study. Nucleic Acids Res., 41:33, 2013.
- M Moradi, V Babin, C Sagui, and C Roland, Recipes for free energy calculations in biomolecular systems. In Biomolecular simulations: methods and protocols. Humana Press (Springer), Methods Mol. Biol., 924:313-37, 2013.
- M Moradi, V Babin, C Roland, and C Sagui, Free energies, structural characteristics, and transition mechanisms of proline-rich peptides. In Proline: Biosynthesis, Regulation and Health Benefits, Nova Publishers, ISBN: 978-1-62257-745-3, 2013.
- M Kalani, A Moradi, M Moradi, E Tajkhorshid, Characterizing a histidine switch controlling pH-dependent conformational changes of the influenza virus hemagglutinin. Biophys. J., 105:993, 2013.
2009-2012
- M Moradi, V Babin, C Roland, T Darden, and C Sagui, Conformations and free energy landscapes of polyproline peptides. Proc. Natl. Aca. Sci. USA, 106:20746, 2009.
- V Babin, V Karpusenka, M Moradi, C Roland, and C Sagui, Adaptively biased molecualr dynamics: An umbrella sampling method with a time-dependent potential. Int. J. Quant. Chem., 109:3666, 2009.
- M Moradi, V Babin, C Roland, and C Sagui, A classical molecular dynamics investigation of the free energy and structure of short polyproline conformers. J. Chem. Phys., 133:125104, 2010.
- M Moradi, J-G Lee, V Babin, C Roland, and C Sagui, Free energy and structure of polyproline peptides: an ab initio and classical molecular dynamics investigation. Int. J. Quant. Chem., 110:2865, 2010.
- M Moradi, V Babin, C Sagui, and C Roland, A statistical analysis of the PPII propensity of amino acid guests in proline-rich peptides. Biophys. J., 100:1083, 2011.
- M Moradi, V Babin, C Sagui, and C Roland, PPII propensity of multiple-guest amino acids in a proline-rich environment. J. Phys. Chem. B, 115:8645, 2011.
- M Moradi, C Sagui, and C Roland, Calculating relative transition rates with driven nonequilibrium simulations. Chem. Phys. Lett., 518:109, 2011.
- M Moradi, V Babin, C Roland, and C Sagui, Are long-range structural correlations behind the aggregation phenomena of polyglutamine diseases? PLoS Comput. Biol., 8:e1002501, 2012.