Md Fulbabu Shaikh
Postdoctoral Research Associate

Fulbabu received his B.Sc. degree in 2014 and M.Sc. degree in 2016 in Physics from Aligarh Muslim University (AMU), India. He then joined the lab of Dr. Parimal Kar at Indian Institute of Technology (IIT) Indore to pursue his doctoral research work in the field of Computational Modeling of Biomolecules and Protein Conformational Dynamics.
After receiving his Ph.D. in Biophysics from IIT Indore, India, he joined Theoretical and Computational Biophysics Group in November 2022 to study the structural and molecular dynamics study of membrane transporters.
Department:
Beckman Institute and Biochemistry
Office Address:
3011 Beckman Institute, 405 N. Mathews Avenue, Urbana, Illinois 61801
Office Phone:
217-904-2487
Email Address:
mfsk@illinois.edu
Personal Web:
shaikhbiophysics
Education
- Ph.D. in Computational Biophysics (2022) - Indian Institute of Technology Indore, India
- M.Sc. in Physics (2016) - Aligarh Muslim University (AMU), India
- B.Sc. in Physics (2014) - Aligarh Muslim University (AMU), India
Research Interests
- Understanding Mechanisms of Biomolecules through the Computational Microscope
- Large-scale Conformational Transitions in Membrane Transporter and Membrane Proteins
- Structural Dynamics and Molecular Mechanisms of Membrane Transporters
- Molecular Modeling and Computer-Aided Drug Discovery
Selected Publications
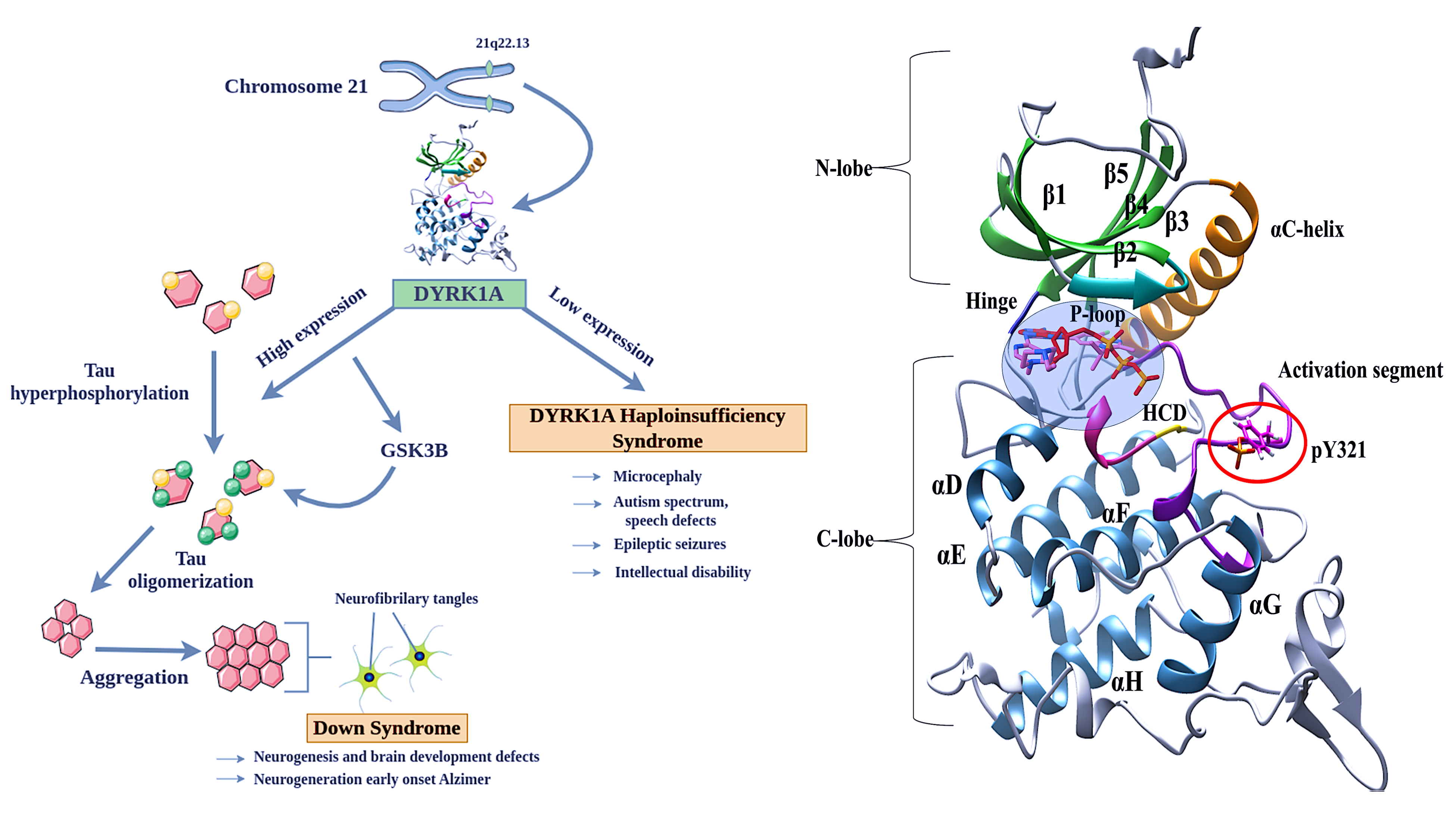
Unlocking the Conformational Secrets of DYRK1A Kinase with Computational Microscope: Exploring Phosphorylation-Driven Structural Dynamics.
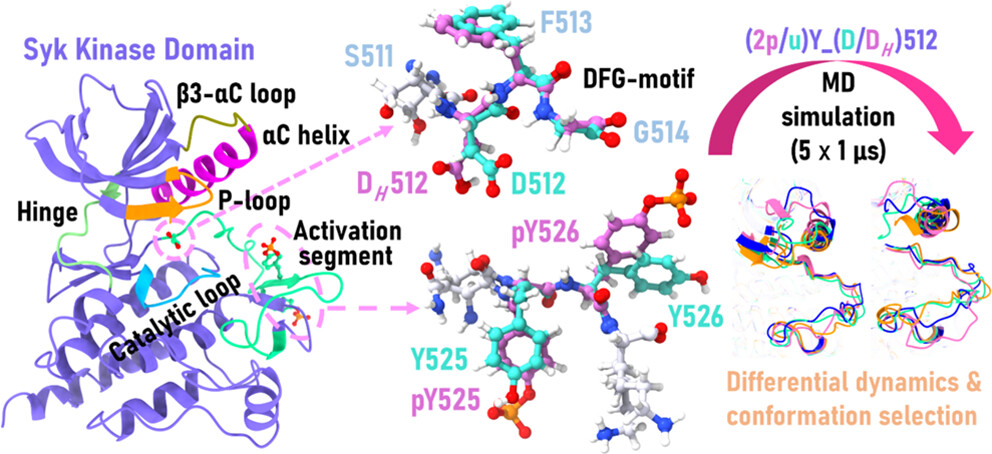
Dynamic Interplay of Loop Motions Governs the Molecular Level Regulatory Dynamics in Spleen Tyrosine Kinase: Insights from Molecular Dynamics Simulations.
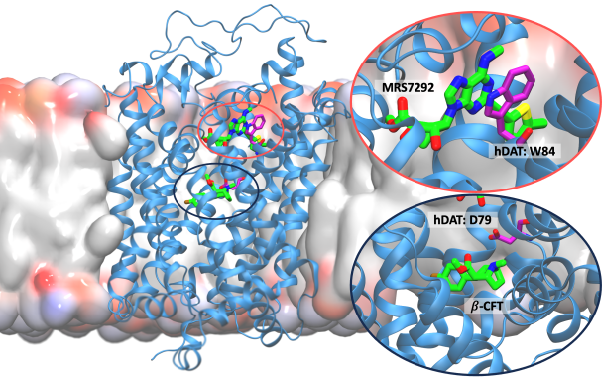
Structure of the human dopamine transporter and mechanisms of allosteric inhibition.
Deciphering the molecular choreography of Janus kinase 2 inhibition via Gaussian accelerated molecular dynamics simulations: A dynamic odyssey.
Microsecond dynamics of H10N7 Influenza Neuraminidase reveals the plasticity of loop regions and drug resistance due to the R292K mutation.
Exploring molecular interactions of potential inhibitors against the Spleen Tyrosine Kinase implicated in Autoimmune disorders.
Computational studies indicated the effectiveness of human metabolites against SARS-Cov-2 main protease.
Phosphorylation-Induced Conformational Dynamics and Inhibition of Janus Kinase 1 by Suppressors of Cytokine Signaling 1.
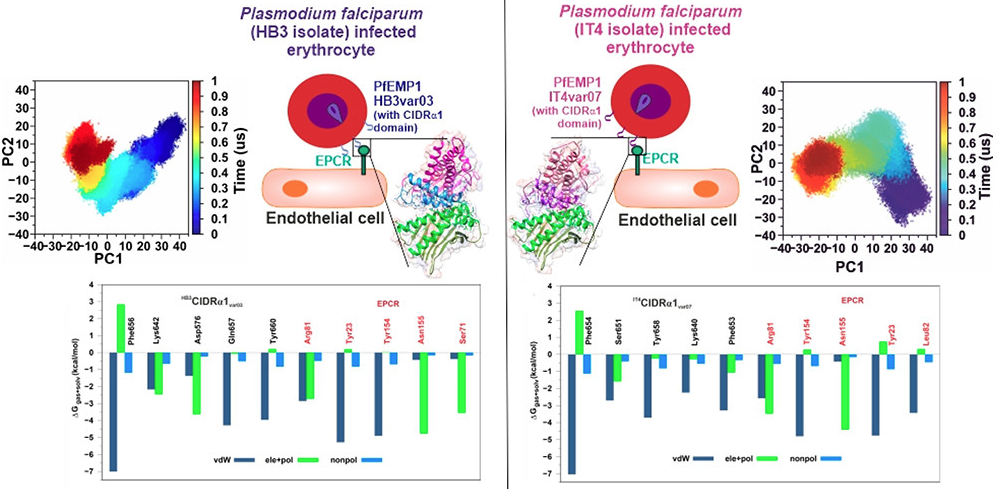
Decoding the Host-Parasite Protein Interactions Involved in Cerebral Malaria Through Glares of Molecular Dynamics Simulations.
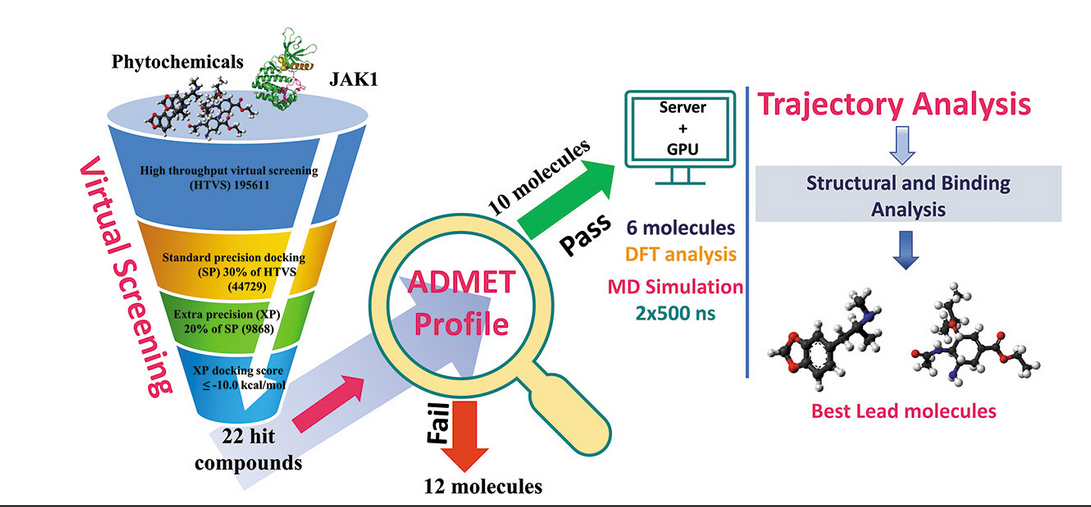
Unraveling the Molecular Mechanism of Recognition of Selected Next-Generation Antirheumatoid Arthritis Inhibitors by Janus Kinase 1.
Finding inhibitors and deciphering inhibitor-induced conformational plasticity in the Janus kinase via multiscale simulations.
A plausible contributor to multiple sclerosis; presentation of antigenic myelin protein epitopes by major histocompatibility complexes.
Characterizing an allosteric inhibitor-induced inactive state in with-no-lysine kinase 1 using Gaussian accelerated molecular dynamics simulations.
Identification of Potential Inhibitors against Epstein-Barr Virus Nuclear Antigen 1 (EBNA1): An Insight from Docking and Molecular Dynamic Simulations.
Unraveling the Molecular Mechanism of Recognition of Human Interferon-Stimulated Gene Product 15 by Coronavirus Papain-Like Proteases: A Multiscale Simulation Study.
Computational investigation of structural dynamics of SARS-CoV-2 methyltransferase-stimulatory factor heterodimer nsp16/nsp10 bound to the cofactor SAM.
Investigating Phosphorylation-Induced Conformational Changes in WNK1 Kinase by Molecular Dynamics Simulations.
#Both contributed equally