Nuclear Hormone Receptors
|
Nuclear hormone receptor proteins form a class of ligand activated proteins that, when bound to specific sequences of DNA serve as on-off switches for transcription within the cell nucleus. These switches control the development and differentiation of skin, bone and behavioral centers in the brain, as well as the continual regulation of reproductive tissues. Researchers at the Theoretical Biophysics Group study the interaction of some members of the nuclear hormone receptor with DNA as well as their interaction with hormones.
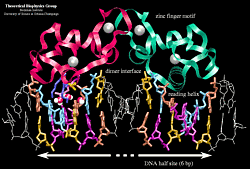
A hormone response element (HRE) is a specific DNA sequence that a receptor recognizes with markedly increased affinity and typically contains two consensus hexameric half-sites. The identity of a response element resides in three features: the sequence of the base pairs in the half-site, the number of base pairs between the half-sites and the relative orientation of the two half-sites. Thus each receptor protein dimer that binds the DNA has to recognize the sequence, spacing and orientation of the half-sites within their response element. The nuclear hormone receptor proteins are composed of several domains which are differentially conserved between the various receptors and have different roles: a variable N-terminal region, a conserved DNA binding domain (DBD), a variable hinge region, a conserved ligand binding domain (LBD), and a variable C-terminal region. DNA Binding Domain (DBD)
How can this flexibility in DNA sequence recognition be correlated with the specific base contacts seen in the crystal structures? Molecular dynamics studies of the binding of the estrogen receptor to specific and non-specific sequences of DNA allowed us to answer this question. Please follow the above link for a detailed description of the study. Ligand Binding Domain (LBD)
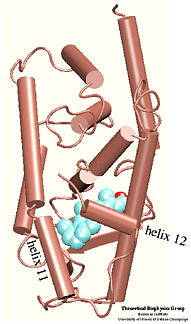
Which residues are most involved in the process of binding/unbinding? How large are the conformational changes induced in the protein by the binding/unbinding process? Can the hormone use one pathway for binding and another pathway for unbinding? These questions were addressed in a study of the unbinding of retinoic acid from its receptor . To induce the unbinding on a time scale accessible to molecular dynamics simulations we used Steered Molecular Dynamics (SMD) simulations. Please follow the above links to find more about the results of this study. |
Publications
|
Dorina Kosztin, Sergei Izrailev, and Klaus Schulten. Biophysical Journal, 76:188--197, 1999. Dorina Kosztin, Thomas C. Bishop, and Klaus Schulten. Biophysical Journal, 73:557--570, 1997. Tom Connor Bishop, Dorina Kosztin, and Klaus Schulten. Biophysical Journal, 72:2056--2067, 1997. |

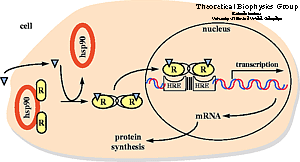 Nuclear hormone receptors are ligand-activated transcription
factors that regulate gene expression by interacting with specific DNA sequences
upstream of their target genes. As early as 1968 a two-step mechanism of action
was proposed for these receptors based upon the observation of an inactive and
an active state of the receptors. The first step involves activation through
binding of the hormone; the second step consists of receptor binding to DNA and
regulation of transcription.
Nuclear hormone receptors are ligand-activated transcription
factors that regulate gene expression by interacting with specific DNA sequences
upstream of their target genes. As early as 1968 a two-step mechanism of action
was proposed for these receptors based upon the observation of an inactive and
an active state of the receptors. The first step involves activation through
binding of the hormone; the second step consists of receptor binding to DNA and
regulation of transcription.