Scientific Content of National Lecture
On 9 February 2015, Klaus Schulten delivered the National Lecture for the Biophysical Society. The topic was "Discoveries in Biophysics Through the Computational Microscope." Our perspective in the Theoretical and Computational Biophysics Group (TCBG) is that the computer is now sophisticated enough in its own right to enable discoveries in biophysics. An overview of the scientific content of the talk is given below. The systems covered provide a broad overview of much of the molecular dynamics research conducted by the TCBG in the last twenty years. The topics discussed, like membrane proteins, mechanical proteins, the HIV virus capsid, and the ribosome, highlight how the computer is being used today to illuminate biological systems. The images come directly from the slides that Schulten used in the National Lecture.
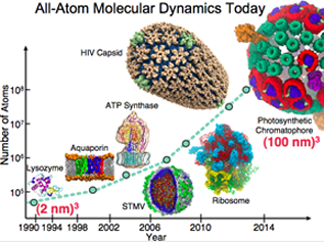 |
Advances in Computational Biology 1990-2015
In molecular dynamics, there has been steady progress in system size obtained. A living cell is made of societies of biomolecules, "and so by going to larger systems we become in many ways more biological." These benchmarks were made possible by our continual research in algorithms, software, and parallel computing.
|
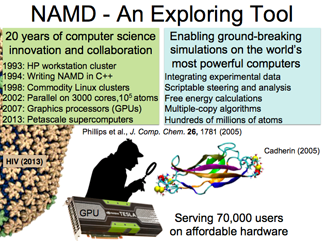 |
NAMD: A Tool to Explore
NAMD is the molecular dynamics simulation program, designed from its outset for parallel computing. It works on the laptop to the supercomputer, and was one of the first programs to utilize GPUs. Developers have constantly transcended limits to make NAMD go faster and larger, and "most of the speed up comes actually from ideas–from mathematics and algorithms." For example, NAMD has incorporated the multilevel summation method to accelerate the notoriously slow Coulomb interaction computation.
MORE
SOFTWARE DEBUT 1996
|
 |
VMD: A Tool to Think
VMD is a visualization and analysis tool that works on small proteins all the way to 100-million-atom structures. The breadth of functionality of this tool is constantly being expanded by plugins, some designed in house, but many contributed by external users, which makes VMD a community effort.
Some plugin examples: Force Field Toolkit, which aids in the laborious process of determining parameters, and Normal Mode Wizard, which calculates and visualizes normal modes.
MORE
SOFTWARE DEBUT 1995
|
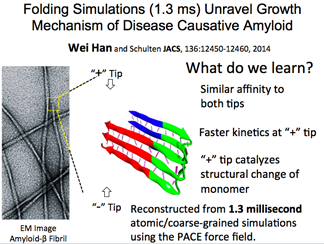 |
Amyloid-Beta Fibril Elongation
Amyloid-beta fibrils are linked to Alzheimer's disease and comprise the plaques seen in patient brains. Now NAMD can simulate elongation of a fibril tip, a very slow process in nature. The innovation, coarse-graining the solvent but describing the peptide at the all-atom level, resulted in over a 1 ms simulation.
We can now see the intermediate states as well as determine the molecular basis for why one tip grows faster than the other.
MORE
DISCOVERY PUBLISHED 2014
|
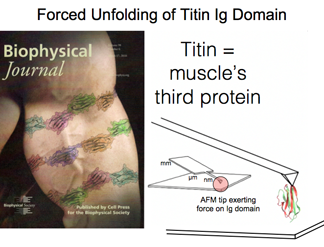 |
Titin: The Mammoth Muscle Protein
Titin, the longest protein of the human genome, plays a key role in muscle elasticity. Although simulation stretched this protein at much faster pulling speeds than in experiment, this early research has recently been validated when experiment and theory finally converged. Over the years computation revealed the molecular nature of the stretching and how numerous hydrogen bonds as well as water molecules play a role in the observed rupture force.
DISCOVERY PUBLISHED 1998
|
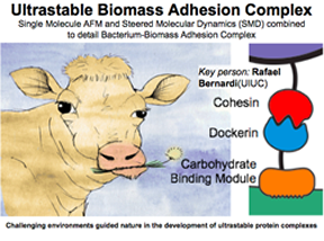 |
Cellulosomes of Gut Bacteria
Bacteria in the rumen of cows help break down cellulose and are being investigated as possible biofuels. On the surface of these bacteria is a tentacle-like system of proteins called a cellulosome, which exists in a constantly moving fluid environment. Simulation has shown how the modular pieces of the cellulosomes grow more adhesive as forces are applied, thereby illuminating the underlying physical mechanism.
DISCOVERY PUBLISHED 2015
|
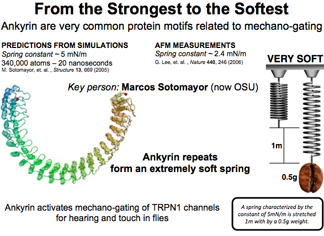 |
Spring Constant of Ankyrin
Ankyrin is a common motif found in scores of human proteins and is believed to be a mechano-gating spring for hearing and touch in flies. Schulten's group raced ahead of experiment and predicted the elastic behavior of ankyrin. What they found was an extremely soft spring, such that a coffee bean could extend it by one meter! Simulation results were shortly confirmed by experimental groups.
DISCOVERY PUBLISHED 2005
|
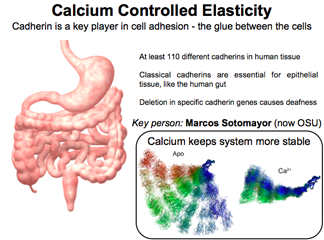 |
Cadherins and the Role of Calcium
Ubiquitous in the human body, cadherins help cells "adhere" (hence their name) to each other. For example, they organize epithelial tissue like that in the human gut. Without cadherins we would just be a soup of cells. While it was known that calcium plays a role for these proteins, simulations revealed that "the elastic rigidity of cadherin is affected by calcium, particularly also the rupture force."
DISCOVERY PUBLISHED 2008
|
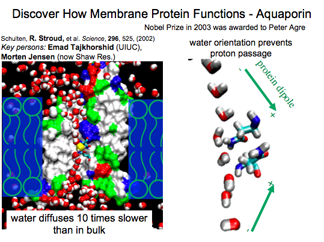 |
Aquaporin and its Water Conduction Demystified
A milestone was reached when computation could embed a protein in its membrane host. Aquaporin conducts water, but how does it exclude protons? Simulations revealed how water molecules change orientation on their trek through the pore, a dynamic process seen only with the computational microscope.
DISCOVERY PUBLISHED 2002
|
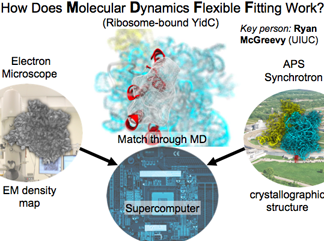 |
Molecular Dynamics Flexible Fitting
This method developed in our group takes high-resolution data (like crystallographic structures) and flexibly fits them into electron density maps.
But the fitting is informed by the "combined wisdom of thirty years of molecular dynamics." MDFF has over seventy reported applications.
METHOD DEBUT 2008
|
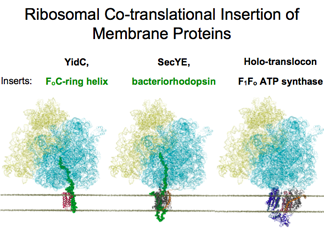 |
Ribosomes and Protein Insertion into Membrane
Studies on the ribosome form a core component of our group. Using MDFF to determine structures, this provides a starting point for further molecular dynamics elucidation. Chaperones guide nascent proteins where they need to go, and our studies examine the dynamic process of integrating proteins into membranes.
DISCOVERIES PUBLISHED 2006-2014
|
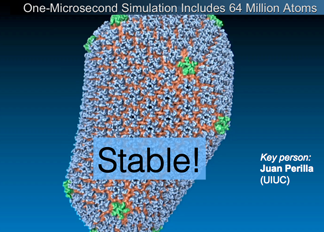 |
HIV Capsid and Chemical Detail
The HIV capsid, the shell that contains the viral genetic material, was structurally determined in full chemical detail, to the tune of 64 million atoms. With every atom known, a clearer picture emerged of the role of the capsid in the infection process
The capsid utilizes "a very intelligent chemistry and communication" on its surface to recruit some cell factors for help and to deter others meant to destroy it.
STRUCTURE PUBLISHED 2012
|
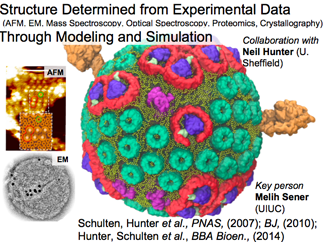 |
Systems Biophysics of Chromatophore
This bacterial pseudo-organelle converts sunlight into ATP. It contains over 100 proteins (6 different types) and we determined a structure of the chromatophore from 40 years of data. Our aim is "to study every process that contributes and interlocks with the other processes in order to understand this system," hence taking a "systems biophysics" approach to unravel how this functional cellular machine operates. More below.
MORE
STRUCTURE DETERMINED 2007
|
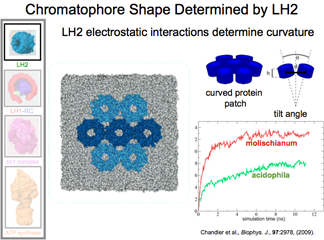 |
LH2 Protein Also Curves Membrane
While a role of the light-harvesting complex 2 (LH2) is to absorb sunlight and then transfer that energy to the reaction center, our studies have also found that it is vital for sculpting the chromatophore into its final spherical shape. This is critical, first, because it allows proteins to find each other more easily, and second, it allows for the right stoichiometry because the number of proteins that fit in a sphere depends on the curvature.
DISCOVERY PUBLISHED 2008
|
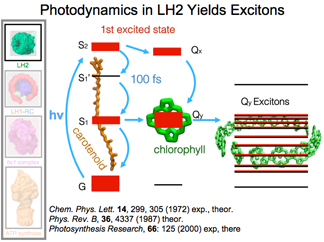 |
Quantum Mechanics at Finite Temperatures
LH2 is made up of rings of chlorophylls and a ring of carotenoids. Once these pigments absorb a photon, this excitation is shared in the ring in the form of an exciton, a coherently delocalized excitation. Even with thermal disorder, there is strong coherency in the ring. Interest is quantum computing at finite temperatures has steered researchers to examine the quantum physics utilized by these organisms at 300K.
DISCOVERY PUBLISHED 2012
|
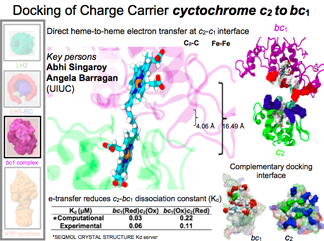 |
Cytochrome c2 and the bc1 complex
In the chromatophore electrons are recycled; the shuttle protein cytochrome
c2 takes them from the bc1 complex back to the reaction center. When cytochrome c2 docks to bc1, a complementary surface is revealed edge-to-edge. "This is nanostructure engineering at its absolute best."
DISCOVERY MADE 2015
|
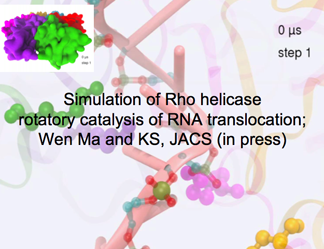 |
The Motor Protein Rho Helicase
This hexameric helicase pumps RNA through its center, powered by ATP. Now all-atom molecular dynamics can reach the relevant time scale of this molecular motor in action, a breakthrough made possible with advanced path sampling techniques. The simulation shows "the intricate organization, coordination and mechanical force delivery that moves the RNA through the system."
DISCOVERY PUBLISHED 2015
|
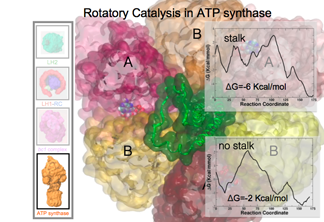 |
ATP Synthase and Rotary Catalysis
ATP synthase is a very famous protein that uses a proton gradient to make molecules of ATP, the cell's currency. Recent breakthroughs (advanced path sampling techniques) have allowed millisecond simulations of this motor protein. Preliminary results so far point to excellent agreement with key intermediate states important in ATP synthesis.
DISCOVERY MADE 2015
|





















