Multi Molecule Animation Plugin, Version 1.0
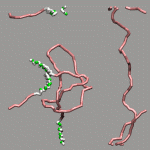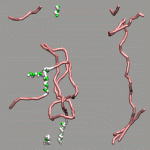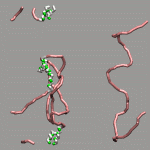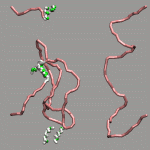
The internal design of VMD does (currently) not support changes of topology and number of atoms during a trajectory for a single 'molecule'. In case of breaking bonds, this can be handled most of the time by using the Dynamic Bonds representation. But if one wants to have a changing number of atoms or display only specific bonds (e.g. as given in a series of psf files), one has to load each into separate 'molecules'. The MultiMolAnim GUI can then be used to emulate the regular animation GUI functionality by selectively displaying only one of the loaded molecules.
To ease the creation of a consistent visualization of all frames/molecules you may want to have a look at the Clone Reps Plugin which allows to transfer visualizations from one molecule to others from a simple GUI.
Graphical Interface:
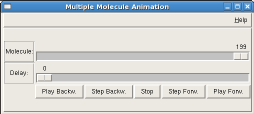
The graphical user interface has two horizontal sliders where you can navigate to an individual frame (='molecule') and set the delay between displaying two consecutive frames/molecules. The set of buttons below allows to run the animation forward or backwards in continuous loops or with single stepping.



