Avidin-Biotin Complex
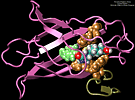
(120K)
MD simulations of protein-ligand adhesion for the avidin-biotin complex.
We performed molecular dynamics simulations which induce, over periods of 40~ps to 500~ps, the unbinding of biotin from avidin by means of external harmonic forces with force constants close to those of AFM cantilevers. The applied forces are sufficiently large to reduce the overall binding energy enough to yield unbinding within the measurement time. Our study complements the work by Grubmueller et al. on biotin-streptavidin which employed a much larger harmonic force constant. The simulations reveal a variety of unbinding pathways, the role of key residues contributing to adhesion as well as the spatial range over which avidin binds biotin. In contrast to Grubmueller et al., the calculated rupture forces exceed by far those observed. We demonstrated, in the framework of models expressed in terms of one-dimensional Langevin equations with a schematic binding potential, the associated Smoluchowski equations, and the theory of first passage times, that picosecond to nanosecond simulation of ligand unbinding requires such strong forces that the resulting protein-ligand motion proceeds far from the thermally activated regime of millisecond AFM experiments, and that simulated unbinding cannot be readily extrapolated to the experimentally observed rupture.



