NAMD 3.0 New Features
New and Improved Methodologies
GPU-resident single-node-per-replicate simulationA new GPU-resident simulation mode is now available to GPU-accelerated multicore and netlrts builds of NAMD that maximizes performance of small- to medium-sized molecular dynamics simulations. In addition to performing force calculations on GPU devices, the GPU-resident mode also performs numerical integration and rigid bond contraints on GPU, with the simulation data residing on the devices between time steps. By eliminating work performed on the CPU and the overhead from per-step transfer of data between CPU host and GPU device, this new mode running on modern GPU hardware provides more than a 2x performance gain over the earlier GPU-offload mode of simulation. The schematic to the right conceptually compares the classic GPU-offload mode timeline to the new GPU-resident one, similar to what an actual profiling reveals. The result of GPU-resident simulation is that the GPU is much more fully utilized, no longer bottlenecked by the CPU. GPU-resident mode supports essential simulation methodologies:
For multi-GPU platforms, GPU-resident mode provides better scaling than GPU-offload due to the cummulative effect on latency of having a CPU bottleneck on multiple devices. Scaling a GPU-resident simulation across multiple GPUs requires a peer-to-peer connection between all pairs of devices. Although this is possible to do over PCIe, good scaling requires a higher bandwidth, lower latency interconnect, like NVIDIA NVLink. Developed by Julio Maia, David Clark, Peng Wang, John Stone, and David Hardy. |
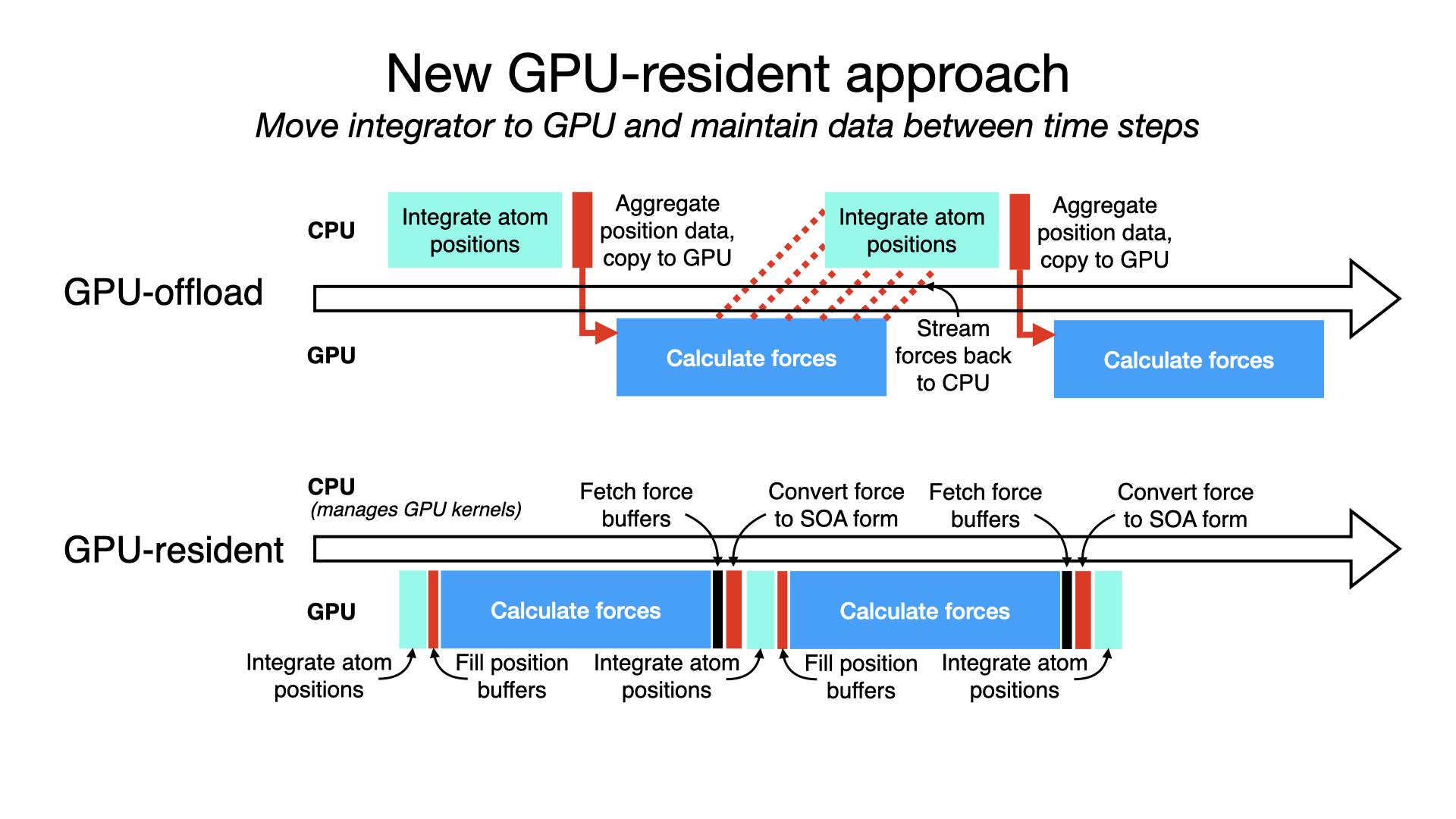
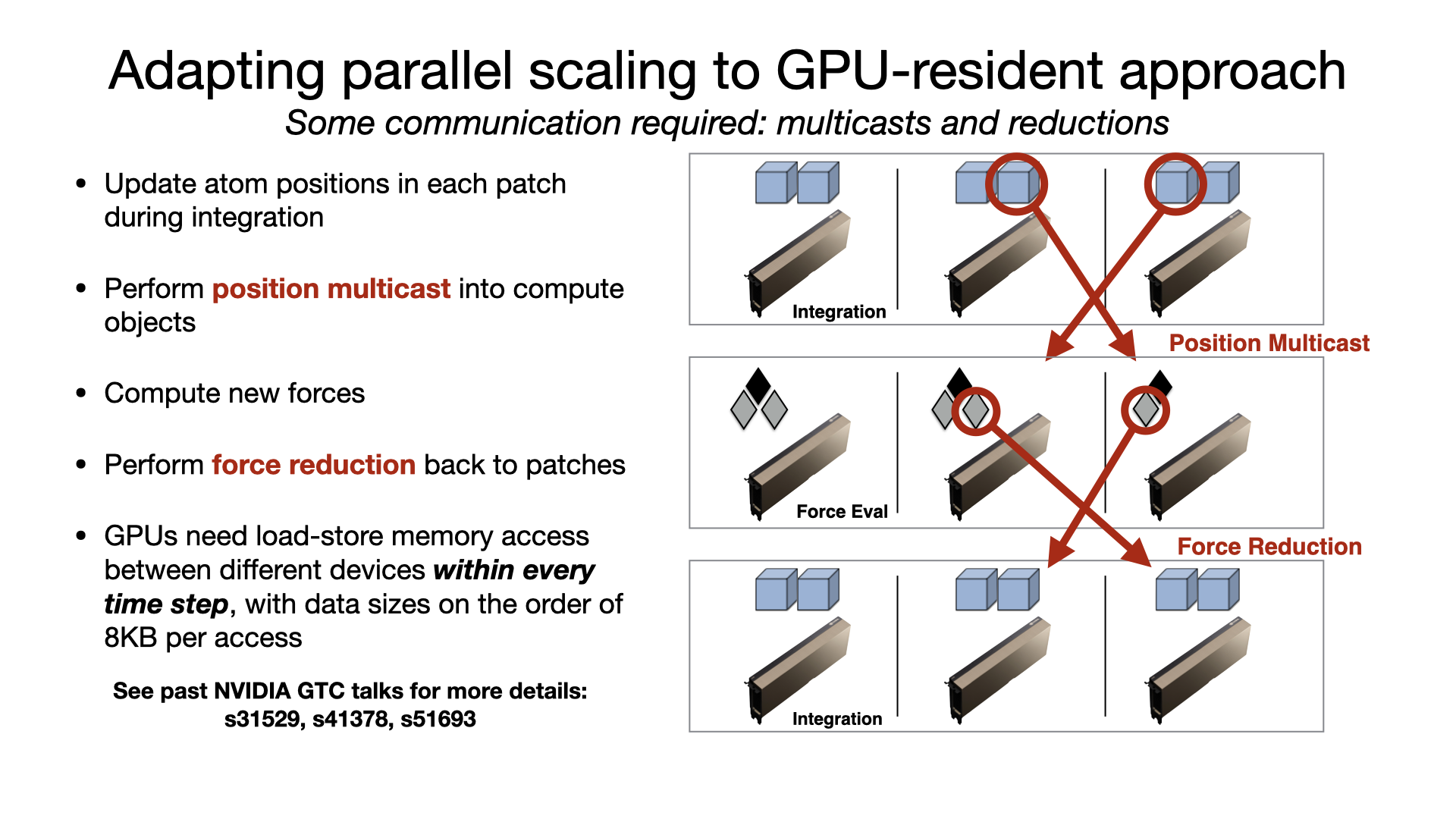
|
GPU-accelerated alchemical free energy methodsThis is the first released version of NAMD to have GPU-accelerated alchemical free energy methods: free energy perturbation (FEP) and thermodynamic integration (TI). These methods calculate free energy differences moving between two different chemical states. Examples include predicting protein-ligand binding affinity and determining solvation free energies. GPU-accelerated support is provided for both GPU-offload and GPU-resident modes, with up to a 30x speedup over the CPU-only implementation in NAMD 2.x. Support is compatible with GPU-resident multi-GPU scaling. Developed by Julio Maia and Haochuan Chen. |
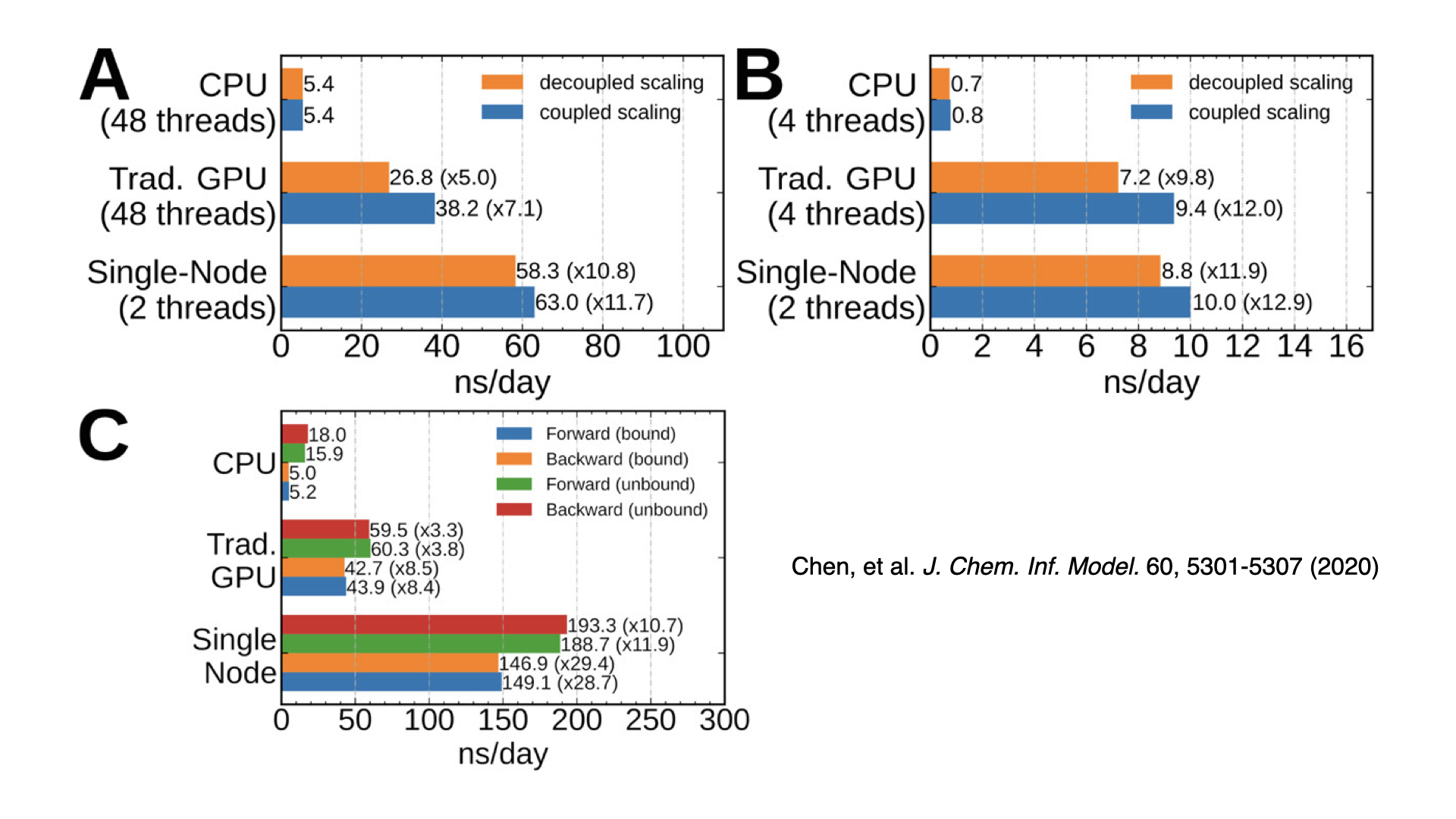
|
Monte Carlo barostat for GPU-resident modePressure is controlled by rescaling the periodic cell at fixed step intervals, and accepting or rejecting this new rescaling based on the Monte Carlo acceptance of the new energy. Since this approach avoids calculating the pressure virial tensor, it is noticeably faster than the Langevin piston barostat. With a rescaling frequency of 100, the MC barostat performance is almost as fast as simulating with thermostat alone. To maintain system stability and achieve a good acceptance ratio, the rescaling needs to be performed on the geometric centers of molecules. Since the communication necessary for a multi-node implementation has not yet been implemented, this feature for now is available only for GPU-resident single-GPU simulation. Developed by Mohammad Soroush Barhaghi. |
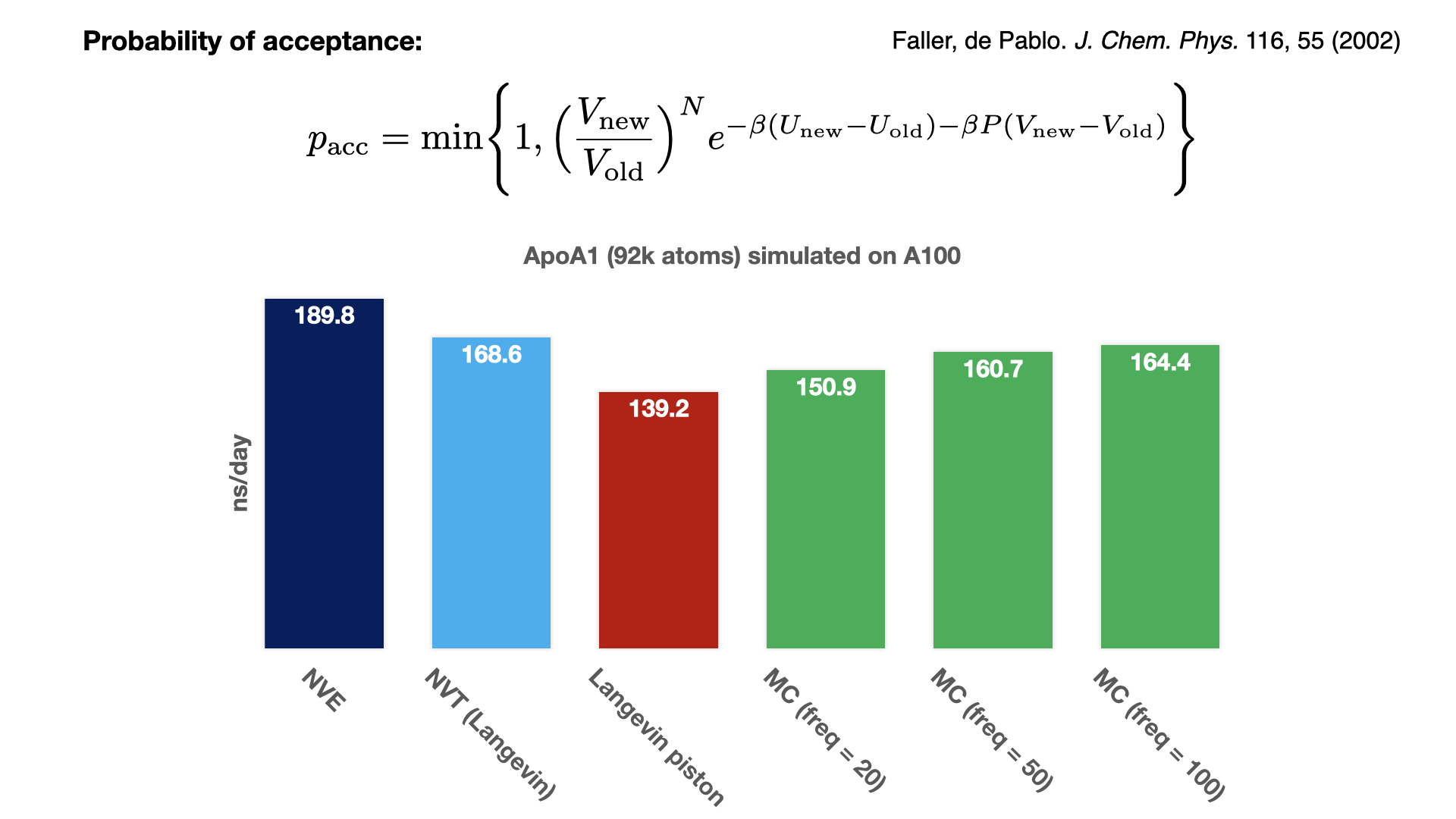
|
Group position restraints for GPU-resident modeThis feature enables a user to define a harmonic restraint either between the centers of mass of two groups of atoms or between the center of mass of a single group of atoms and a fixed point in space. More than one such restraint can be defined. The group position restraints feature provides native support for a common collective variable use case with the Colvars module, giving an indication of the performance possible from a custom implementation designed directly for GPU-resident mode simulation. Comparing performance for a membrane system, restraining the top and bottom halves of a lipid bilayer during equilibration, shows that running GPU-resident with group position restraints is almost 3x faster than running GPU-offload with Colvars, an even bigger improvement than when comparing performances of the unrestrained system. Developed by Mohammad Soroush Barhaghi. |
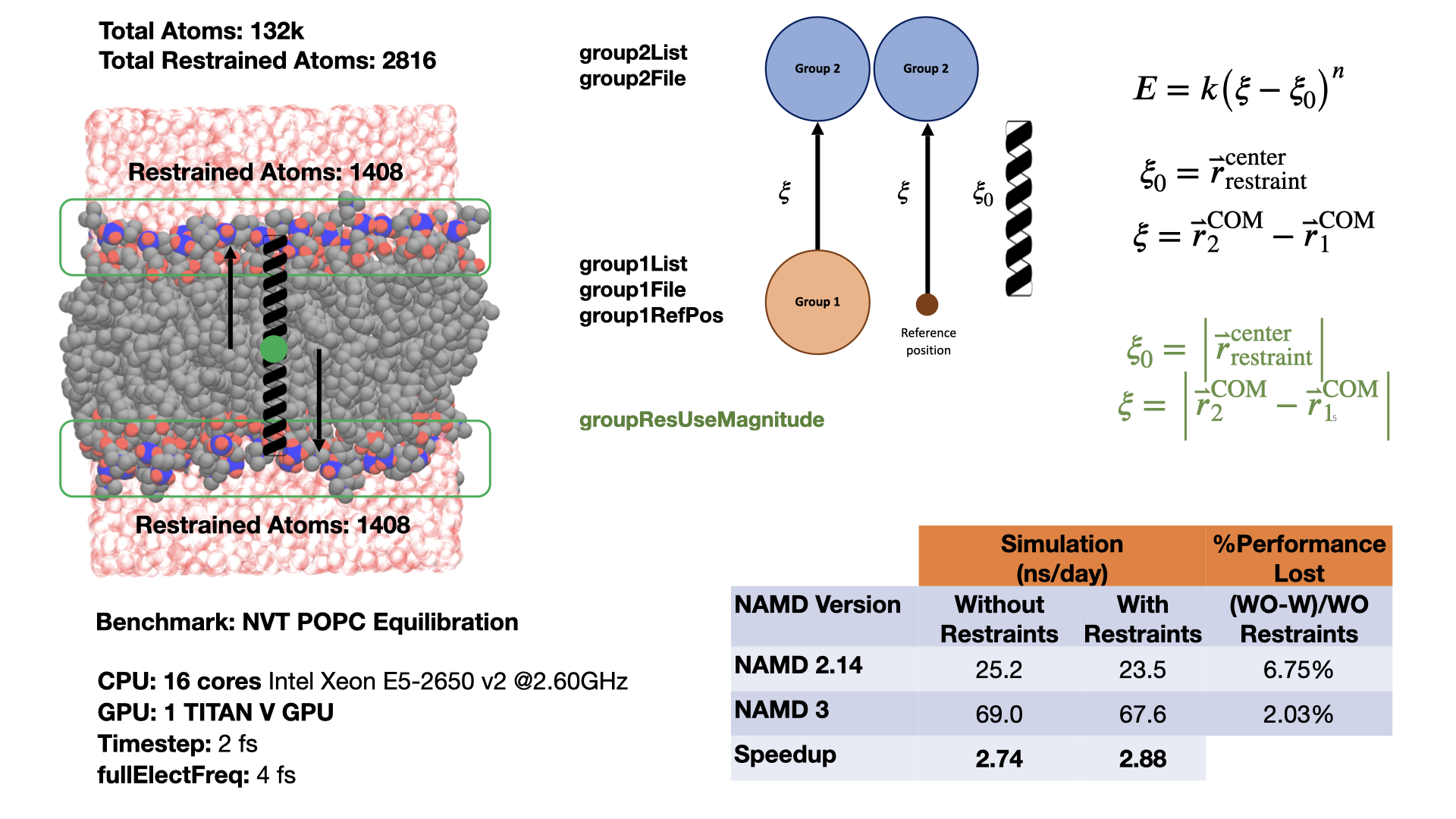
|
Additional improvementsNAMD has the following additional improvements:
|
New Platforms and Scalability
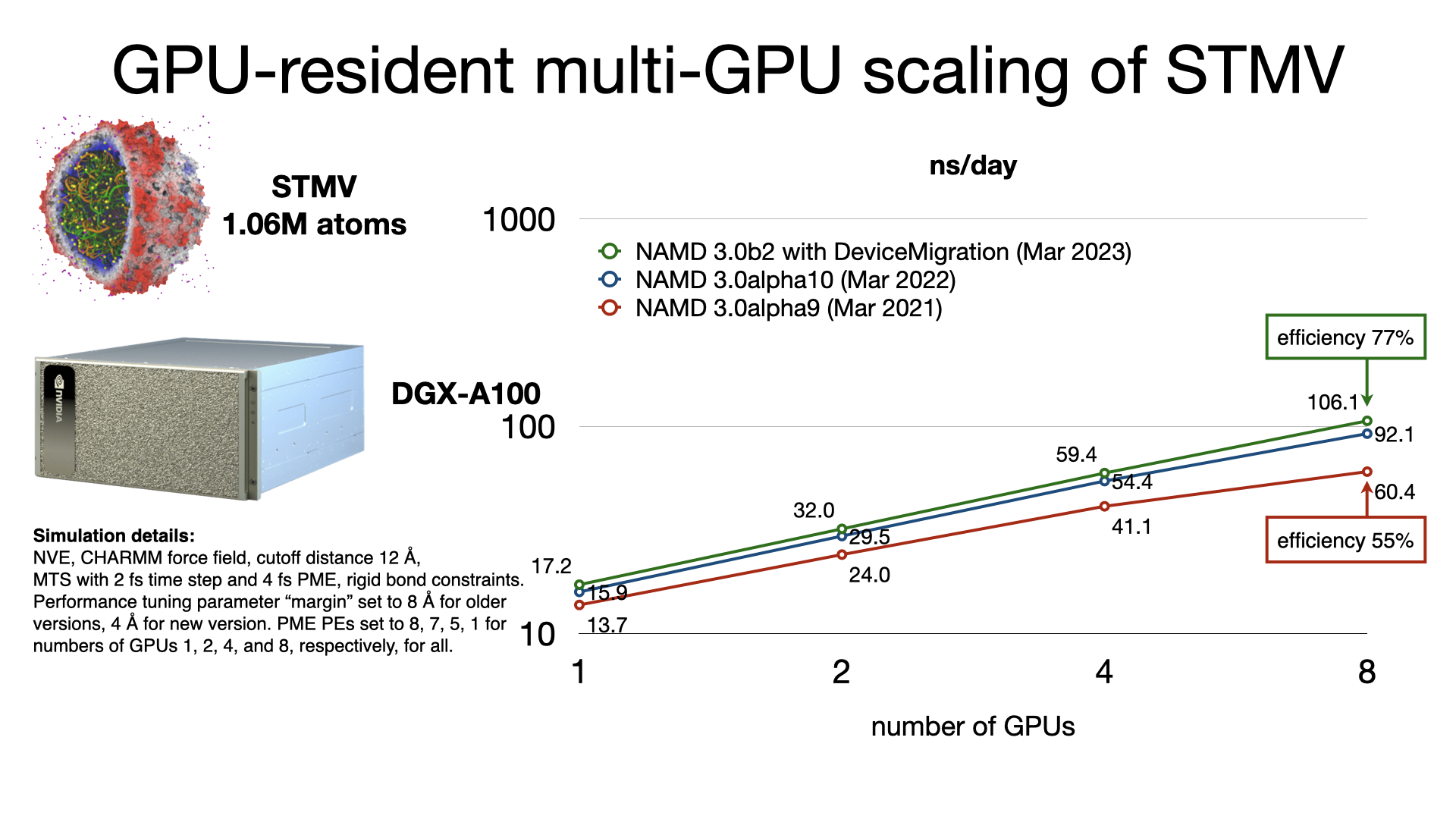
GPU-resident single-node multi-GPU scalingThe released multicore and netlrts builds of NAMD are capable of running GPU-resident mode simulations with multi-GPU scaling on supported platforms. GPU-resident simulation is enabled by setting the configuration parameter:
The benchmarks shown here used appropriate
settings for +pmepes to load balance
the PME computation and also used the new experimental
DeviceMigration feature that performs
atom migration on the GPU devices.
|
|
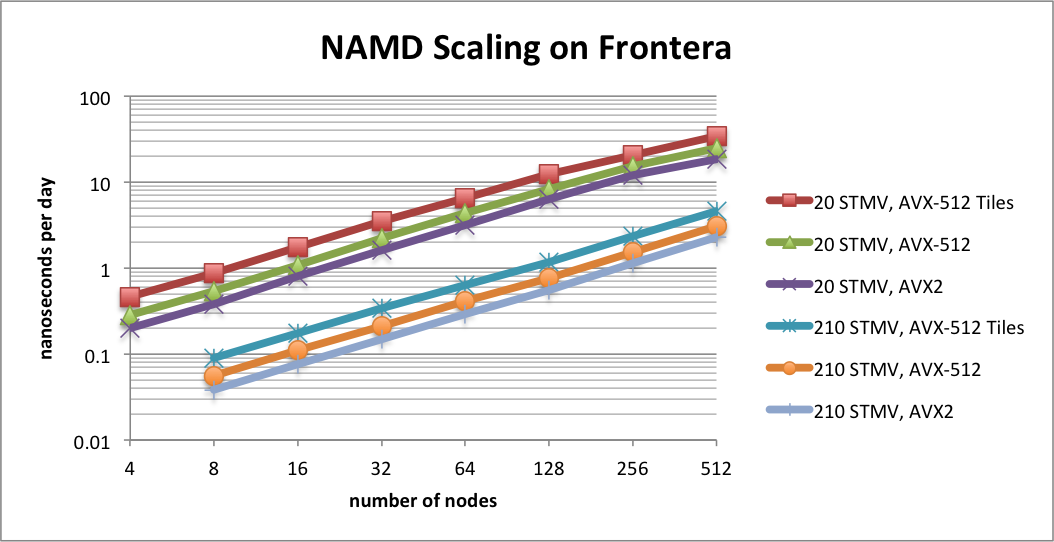
Scaling on the Frontera supercomputerTACC Frontera is the leading NSF computing resource, ranked as the 19th fastest supercomputer on the Top 500 List. NAMD now has special support for Frontera's Intel Cascade Lake CPUs using a new AVX-512 tiles optimization. This optimization provides up to a 1.8x speedup for CPU-based simulations on AVX-512 capable hardware, such as Intel Xeon or AMD Zen4. Developed by Mike Brown (Intel). |
|



