Examples of Past Collaborations
The Center for Macromolecular Modeling and Bioninformatics (CMMB) has engaged in productive and successful collaborations with scientists at local, national, and international locations. A sample of those collaborations is provided below. Visitors may also wish to search our publications database for collaborative articles related to their own area(s) of expertise.
Local
Translocation of DNA by Molecular Motor Helicases
|
Collaborators: Taekjip Ha, University of Illinois at Urbana-Champaign
|
Systems:
- Non-hexameric helicases: UvrD, PcrA, NS3
- Hexameric helicases: E1, DnaB & T7
Importance:
- Do subunits translocate by collective or independent action?
- What is the chemomechanical coupling that links ATP hydrolysis to conformational changes that control translocation?
Challenges and our expertise:
- Modeling by QM/MM, free energy methods & accelerated MD
- Bridging multiple scales, from femtoseconds to milliseconds
- Developing atomic-based models of chemomechanical coupling and energy transduction to describe how ATP hydrolysis powers translocation.
|
Monomer subunits in a hexamer helicase
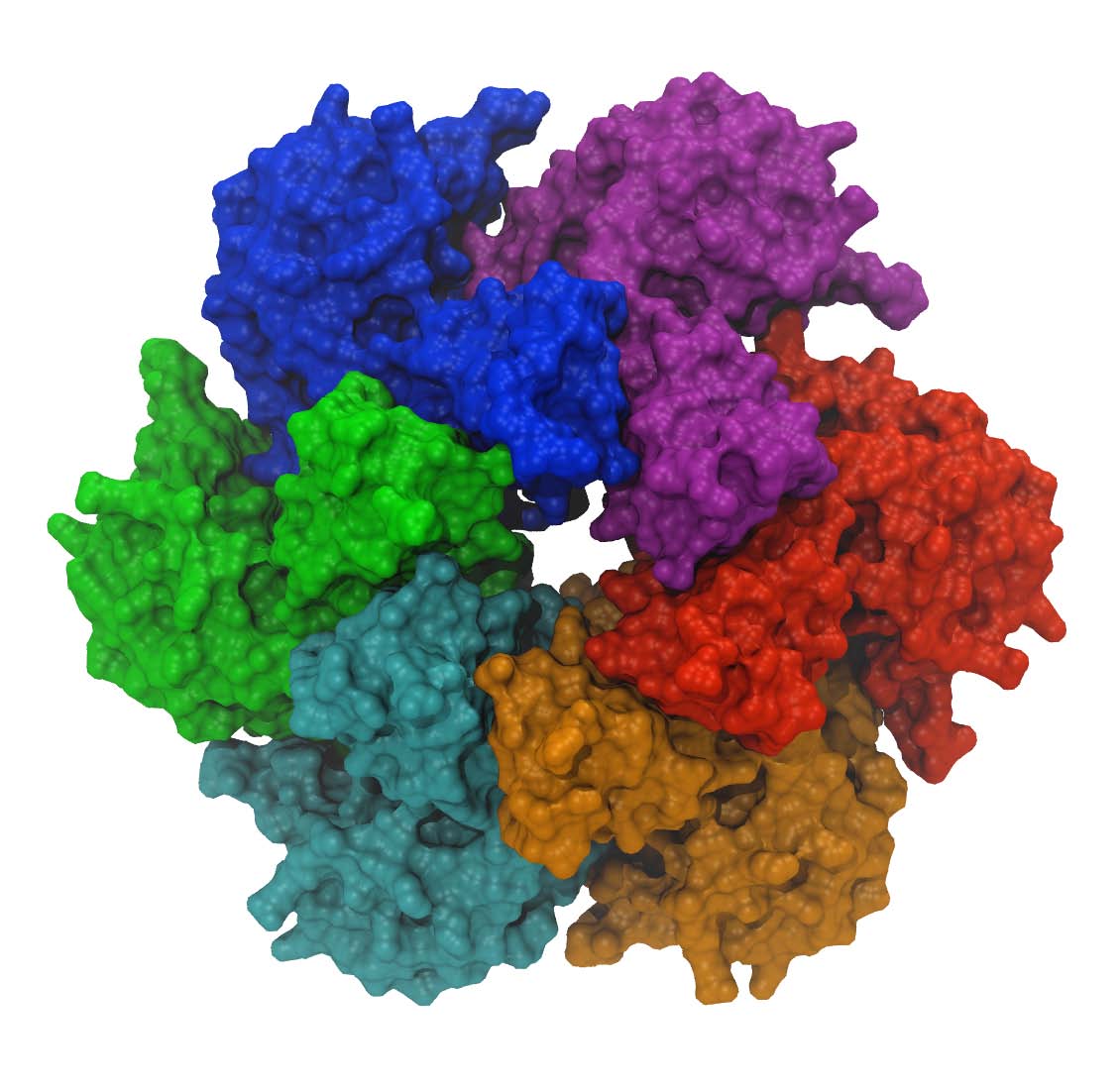
|
Related paper: Leonardo G. Trabuco, Eduard Schreiner, John Eargle, Peter Cornish, Taekjip Ha, Zaida Luthey-Schulten, and Klaus Schulten. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. Journal of Molecular Biology, 402:741-760, 2010. (PMC: 2967302)
|
National
Ribosome: Decoding of Genetic Information
|
Collaborators: Joachim Frank, Howard Hughes Medical Institute, Columbia University
|
System:
- Entire ribosome (~3M atoms); cryo-EM and X-ray data.
Importance:
- Universal machine for translating genetic information into proteins.
- Decoding is target of a number of antibiotics.
Challenges and our expertise:
- How does the ribosome ensure correct decoding of the genetic message?
- Combine cryo-EM and X-ray data.
- MD simulations of large systems (up to 3M atoms).
- Long simulations (>100 ns).
- Expertise in modeling.
|
Ribosome
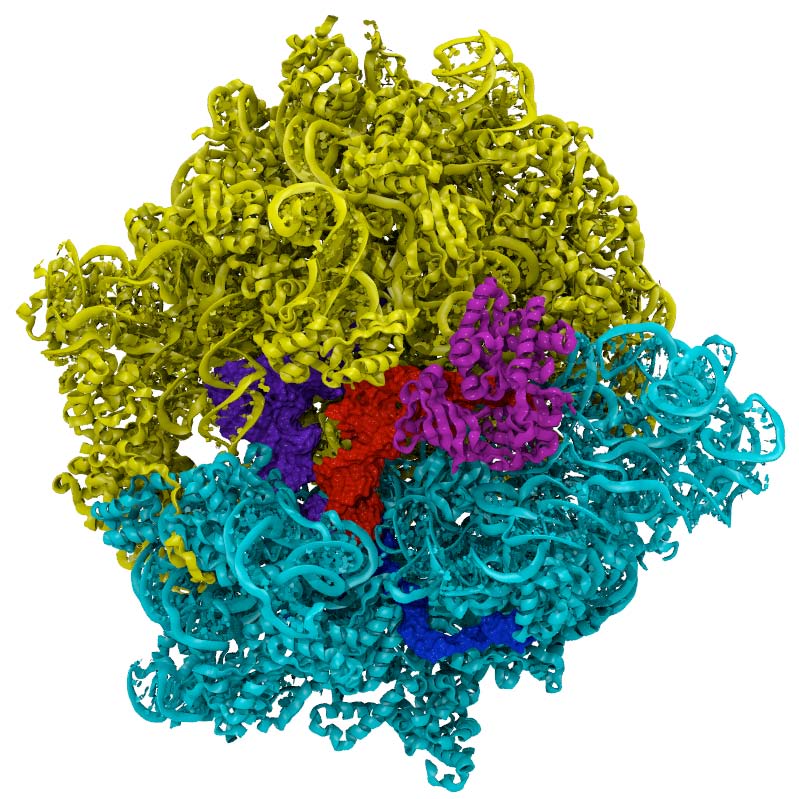
|
Related paper: Xabier Agirrezabala, Eduard Scheiner, Leonardo G. Trabuco, Jianlin Lei, Rodrigo F. Ortiz-Meoz, Klaus Schulten, Rachel Green, and Joachim Frank. Structural insights into cognate vs. near-cognate discrimination during decoding. EMBO Journal, 30:1497-1507, 2011
|
International
Structurally Resolved Photosynthetic Membrane
|
Collaborators: Simon Scheuring, Institut Curie
|
System:
- Photosynthetic bacterial membrane; structural model resolved by atomic force microscopy.
- Membrane patch containing 7 LH1s and 36 LH2s, ~ 25 million atoms.
Importance:
- Photosynthetic bacterial membrane provides a simple model to understand photosynthetic structures.
Challenges and our expertise:
- Characterize interactions of hundreds of LH1 and LH2 proteins in formation of various chromatophore shapes in bacterial membrane.
- Investigate migration of quinones through densely-packed proteins to reach the bc1 complex.
- New versions of VMD and NAMD capable of handling large (25M+ atoms) systems.
- Access to the Center's considerable computational facilities.
|
All-atom representation of lamellar chromatophore patch
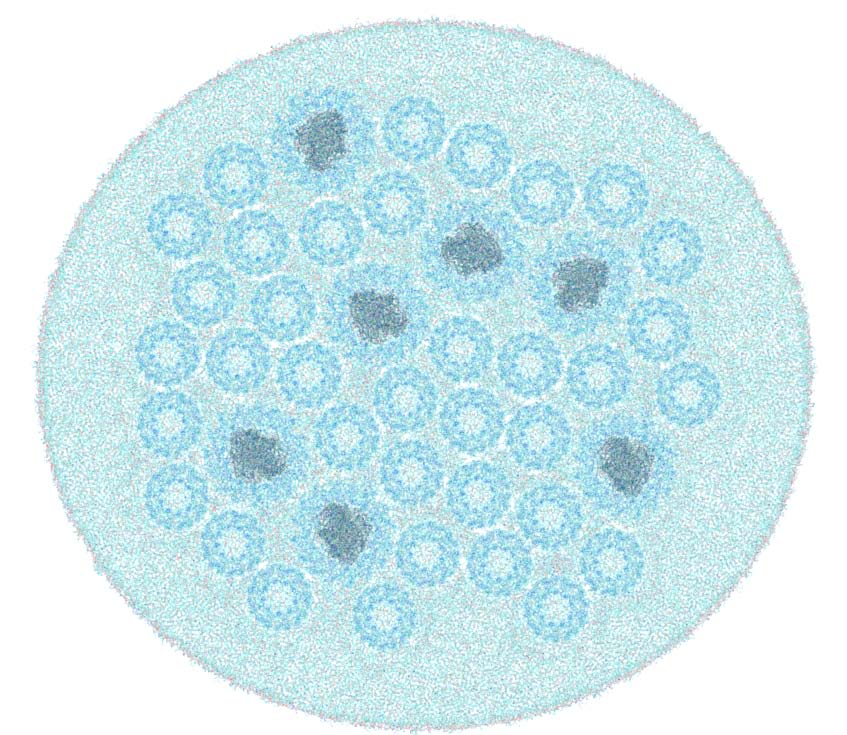
|
Related paper: Melih Sener, Johan Strümpfer, Jen Hsin, Danielle Chandler, Simon Scheuring, C. Neil Hunter, and Klaus Schulten. Förster energy transfer theory as reflected in the structures of photosynthetic light harvesting systems. ChemPhysChem, 12:518-531, 2011. (NIHMS: 285267)
|






