Highlights of our Work
2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001

Click to view the full-size image
Image size:
1.6MB
Made with VMD
The human hERG potassium channel is essential for maintaining normal cardiac rhythm. Mutations of hERG, particularly in its voltage‑sensing arginines, can lead to long‑QT syndrome, a condition characterized by potentially fatal arrhythmias. However, the structural basis of how these mutations impair the voltage sensing of the channel has remained elusive. In our recent Nature Communications study, we combined imaging and single‑molecule experiments with molecular dynamics simulations to capture the intermediate conformations of the voltage‑sensing domains. Simulations were visualized and analyzed using VMD, while simulations were run in NAMD. Our results reveal that disease‑linked mutations alter charge transfer within the sensor, hindering movement and thereby disrupting channel gating. These insights help explain the molecular basis of arrhythmias and suggest strategies to restore hERG channel function.
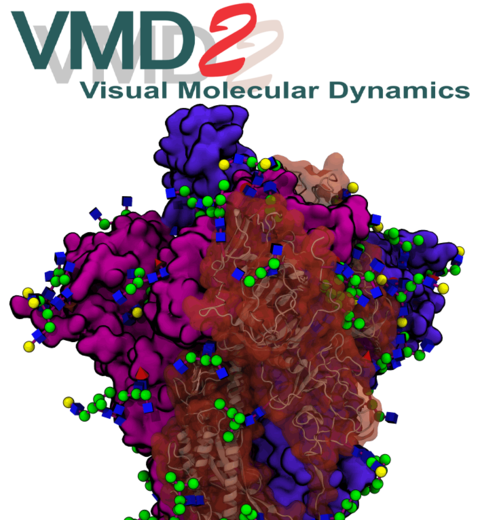
Anion Exchanger 1 (AE1) is the most abundant membrane protein in human red blood cells and the fastest known transporter. It plays a central role in facilitating carbon dioxide transport from tissues to the lungs by rapidly exchanging chloride and bicarbonate ions across the membrane. In collaboration with the Clarke Lab at Columbia University and using molecular dynamics simulations and advanced enhanced-sampling techniques with NAMD, the Resource researchers revealed the mechanism by which depletion of the anionic lipid PIP2 significantly lowers the rate of the transporter. Analysis of the simulations in VMD uncovered that the formation of a salt bridge between the lipid and AE1 residues modulates the transporter's structural dynamics and function. More details can be found in our publication in Nature Communications.

image size:
796.9KB
provided by Molywood Team
Molywood, the popular tool for producing molecular movies, is now fully integrated in VMD 2.0 (Linux and macOS releases). The new Molywood plugin gives VMD users the ability to create advanced videos directly within VMD, eliminating the need for external scripting. Users can easily combine dynamic highlighting of different molecular regions, dynamic plots, trajectory animations, and smooth camera transitions. The visualization capabilities of Molywood are documented through its library of samples and a growing list of video tutorials. The integration of Molywood in VMD 2.0 greatly facilitates creation of publication-quality molecular animations, allowing researchers to focus on scientific storytelling rather than technical implementations. The tool supports the full spectrum of molecular visualization needs, from simple trajectory playback to sophisticated multi-panel presentations. Advanced users of Molywood can customize and automate comparative analysis of trajectories and ensembles.
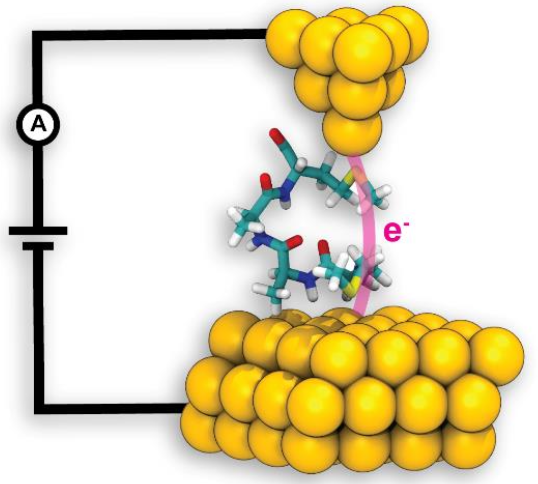
image size:
3.1MB
made with VMD
Proteins have long been involved in biological electron transport, one of the most fundamental reactions in nature. Understanding the link between the structure and electron transport properties of proteins is of utmost importance when using them as templates for synthetic systems. In close collaboration with single-molecule experiments, Resource researchers performed simulations using NAMD to understand the role of secondary structure in electron transport. The results, published in a recent paper in PNAS, highlight the importance of helical conformations in electron transport of peptides and provide broad avenues for understanding the electronic properties of proteins.

image size:
335.7KB
provided by Workshop Organizers
The 63rd Hands-on Workshop on Computational Biophysics, taking place from 8/4 to 8/8 at Auburn University, is the largest workshop we have hosted in recent years! We were able to accommodate more than 60 participants selected from more than 300 applicants. The workshop covered a multitude of topics, from the basics of molecular dynamics to advanced free energy calculation methods, plus the tricks and techniques of visualizing and animating molecular structures using the newest VMD 2.0! We have received highly positive feedback in the post-workshop survey. Stay tuned for the future workshop announcements later this year! You can read more about the workshop's broader impact in this Auburn University news release.

image size:
465.7KB
made with VMD
β2-glycoprotein I (β2GPI), a key autoantigen in antiphospholipid syndrome (APS), binds to negative lipids exposed on damaged cells, thereby enhancing its recognition by pathogenic antibodies. This membrane association—mediated by its fifth domain (DV)—is critical for disease progression, yet its molecular basis remains unresolved. As highlighted in our recent publication in J. Thromb. Haemost., Resource researchers employed molecular dynamics simulations using NAMD to investigate this key membrane binding event. Using an accelerated membrane model developed previously by the Resource, they captured membrane binding of DV and revealed how distinct interactions orchestrate its insertion into negative membranes. The study provides the first atomic-level insight into β2GPI’s membrane association and establishes a structural framework for novel therapeutics in APS.
We are proud to announce our 63rd Hands-on Workshop on Computational Biophysics this summer.
Dates: August 4 - 8, 2025
Location: Leach Science Center, Auburn University
Application Deadline: June 20, 2025
The workshop is ideal for students and researchers from computational and biophysical backgrounds; non-specialists eager to learn computational biophysics methods are also welcome to apply. Topics include molecular dynamics simulations using NAMD 3.0, biomolecular visualization and analysis using VMD 2.0, and advanced modeling, simulation, and analysis methods utilizing NAMD and VMD Plug-Ins.
Cholesterol, an important fat molecule, regulates the structure and function of many membrane proteins. Computational studies of protein-cholesterol interactions have been long impeded by the slow dynamics of lipids like cholesterol. In a recent paper, Resource researchers report a new generation of an accelerated membrane model (HMMM) in which cholesterol can be included. Utilizing NAMD and in-house Tcl scripts with VMD, the new model successfully identifies cholesterol binding sites on proteins and captures detailed lipid interactions in agreement with experimental data.

File size:
40.5MB
Made with VMD
See also additional movie:
27.1MB
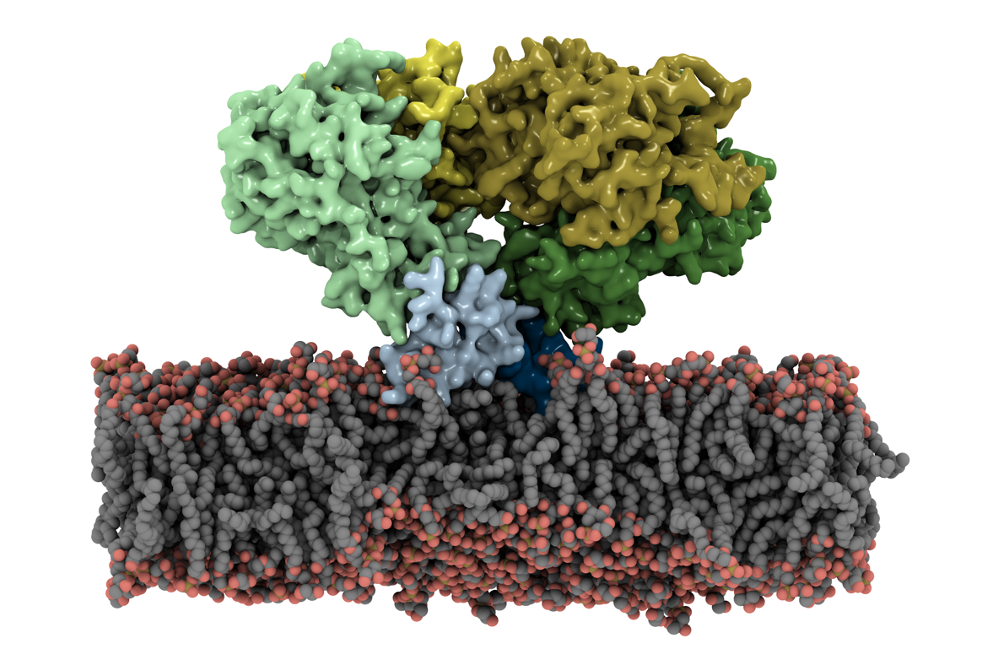
Glutamate transporters maintain brain health by clearing the main excitatory neurotransmitter, glutamate, from synapses. Their dysfunction is linked to neurological conditions such as Alzheimer and Parkinson. In a recent publication in PNAS, Resource researchers characterized the most detailed view of operation of a bacterial glutamate transporters using advanced molecular dynamics simulations with NAMD. The study revealed a sophisticated "elevator-like" mechanism involving both translational and rotational shifts within the protein. Researchers also demonstrated how surrounding lipids influence transporter function. The results enhance our understanding of neurotransmitter regulation, aiding in the design of new therapies for neurological disorders.
As a part of the 2025 Beckman Institute Open House (BIOH), the Theoretical and Computational Biophysics Group (TCBG) offered an exciting and immersive glimpse into the molecular world of modern biology through cutting-edge virtual reality (VR) experiences. As developers of the widely used molecular simulation tools NAMD and VMD, TCBG takes pride in making science accessible to and inspiring for the broader community. At this year's BIOH, visitors had the opportunity to fly through molecular structures, manipulate DNA interactively, and step inside protein complexes that power living cells, all in VR. A standout attraction was the Minecraft-style virtual cell exploration, where guests navigated through blocky but biologically accurate cellular environments generated from real 3D microscopy data. The event drew curious minds of all ages, from young aspiring students to seasoned researchers, showcasing TCBG's ongoing mission to bridge science, technology, and public engagement.
VMD 2.0 is here! This early alpha release marks a significant upgrade to the widely used molecular visualization and analysis software, introducing a streamlined interface, improved selection tools, and cutting-edge performance enhancements. Further development is ongoing, with new features continuously added. Learn more and follow the progress here: VMD 2.0 Introduction.
Major feature enhancements in VMD 2.0 include:
• A redesigned user interface providing a more intuitive and efficient experience; new action buttons provide quick access to basic functions, while an improved selection engine allows for precise molecular filtering.
• Fast & scalable secondary structure calculation
• High-performance glycan visualization
• Rapid & efficient surface calculation
• Real-time interactive ray tracing
With these enhancements and more on the way, VMD 2.0 is shaping the future of molecular visualization. Stay updated and try the latest release: VMD 2.0 Introduction.
Staphylococcus aureus, a prominent human pathogen, employs a specific enzyme to evade the immune system. This enzyme converts antimicrobial fatty acids in the cellular membrane into anti-inflammatory compounds that protect the pathogen against the host inflammatory response. Understanding the membrane-binding mechanism of this enzyme is essential for developing new strategies to combat S. aureus infections.
As highlighted in our recent publication in J. Biol. Chem., Resource researchers employed molecular dynamics simulations using NAMD to study this mechanism. Utilizing a specifically designed membrane model with enhanced lipid motion, they captured how the enzyme binds to the membrane and described its pose within the membrane. These findings provide molecular insights into S. aureus' virulence and highlight potential targets for novel therapeutic interventions.
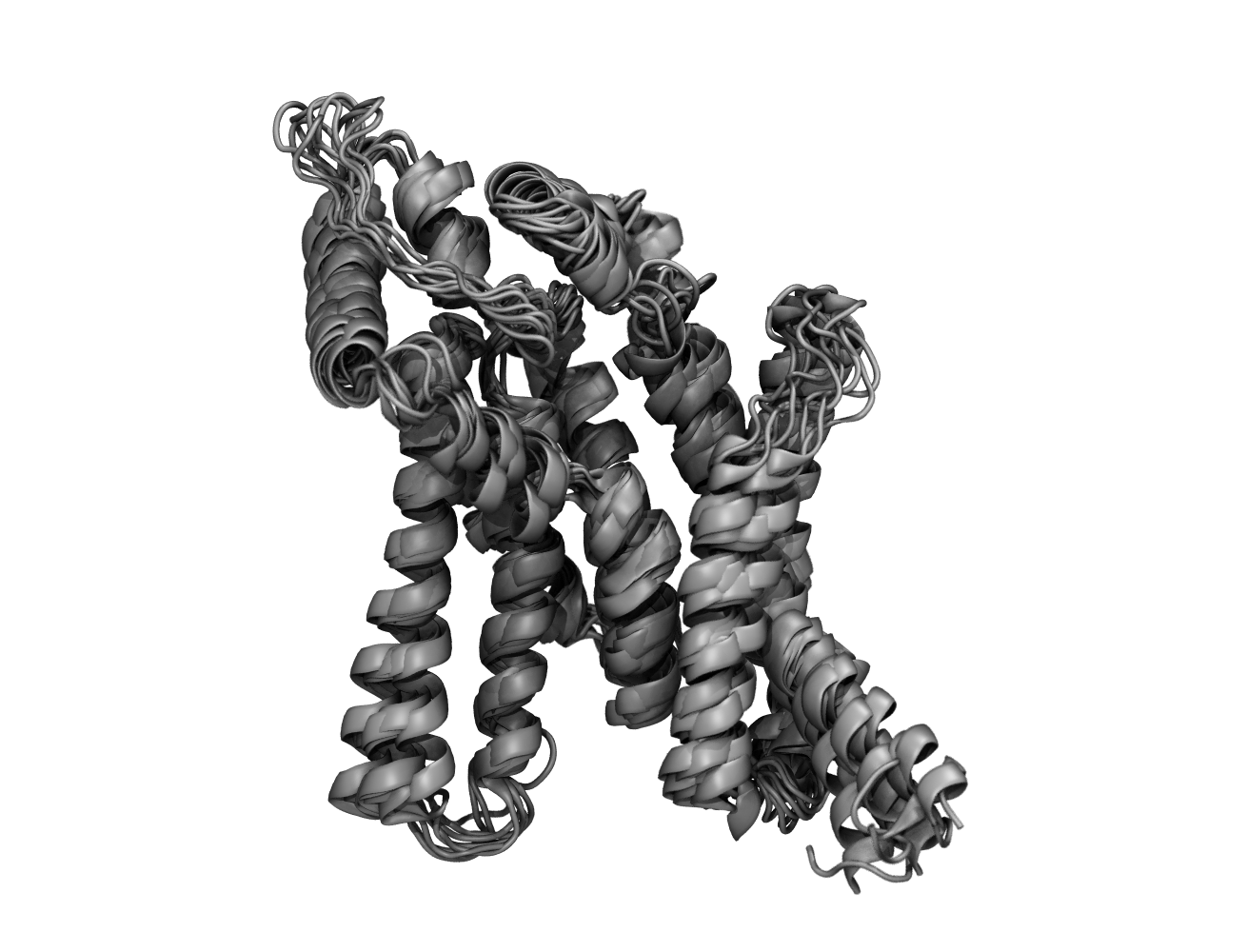
Nearly 80% of the patients suffering from glycogen storage disease have a single mutation in their genome. The product of this gene mediates critical steps in sugar metabolism and storage. However, the molecular basis for the malfunction caused by this and similar mutations has remained unknown due to a lack of structural data. The advent of AlphaFold provided us with the first model of this important protein. As reported in a recent publication in PNAS Nexus, Resource researchers have produced large conformational ensembles of this protein in the empty and sugar-bound forms for both native and 10 clinically-relevant mutants. By identifying the structural and energetic effects of such mutations, using NAMD and VMD, we have begun to elucidate the role of each residue in the dysfunction of the enzyme.