MDFF News and Announcements
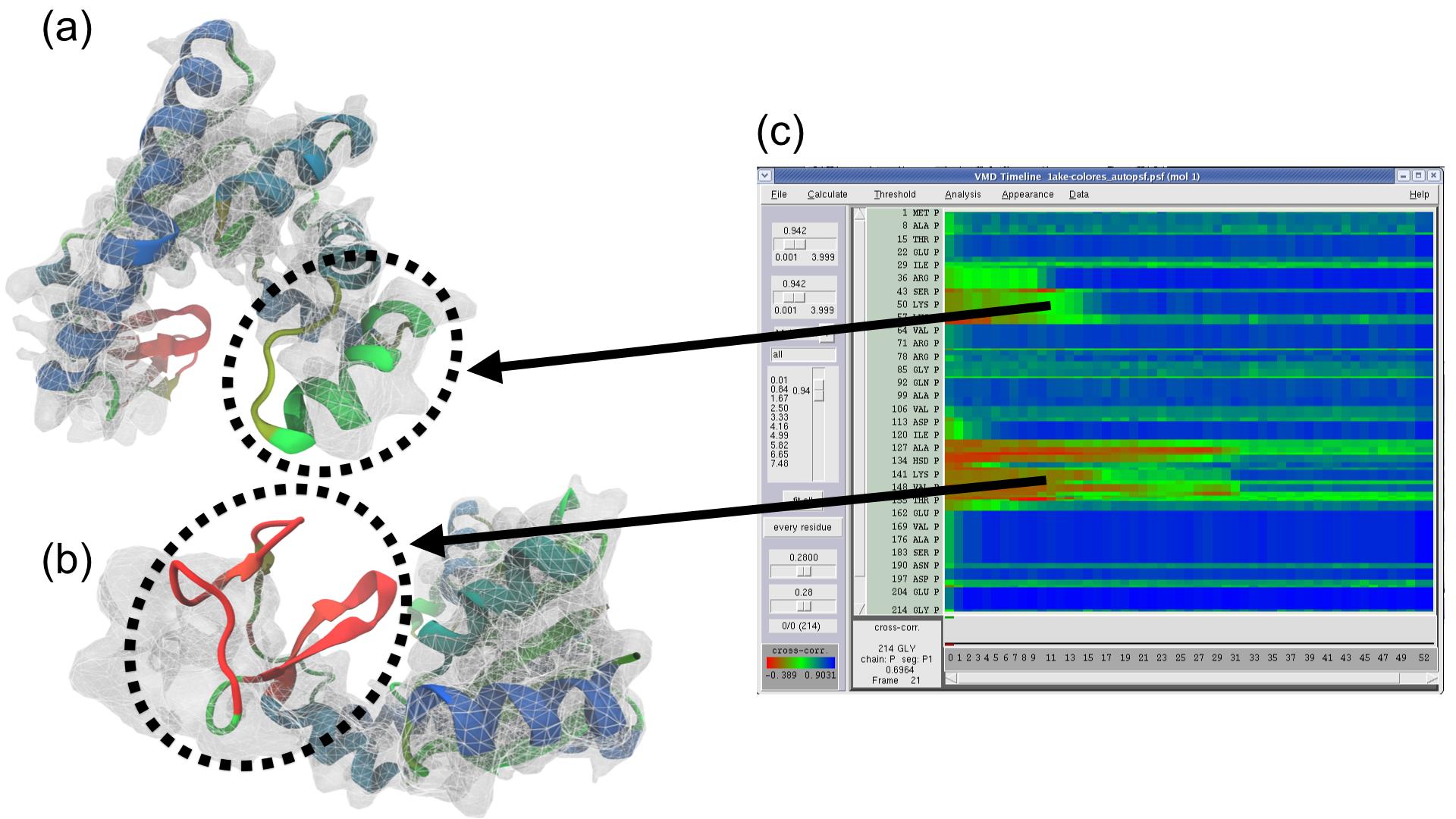
February UPDATE: The MDFF tutorial has now been updated to cover the latest MDFF features outlined below, including the MDFF GUI, interactive MDFF, and Timeline cross correlation analysis. VMD 1.9.2 contains several updates for the Molecular Dynamics Flexible Fitting (MDFF) Method. The mdff plugin contains new options for setting up MDFF simulations with implicit solvent and the new MDFF method for low-resolution x-ray crystallography (xMDFF, recently detailed in this article). Complete details for setting up xMDFF simulations with this plugin can be found in the MDFF tutorial. Additionally, a new graphical user interface for the mdff plugin is now available in the Modeling section of the Extensions menu. This interface allows for easier setup of MDFF and xMDFF simulations, as well as running, connecting to, and analyzing interactive MD simulations. VMD 1.9.2 also contains new multi-core CPU and GPU-accelerated analysis capabilities for MDFF, as described in a recent article. The mdffi cc command can be used to quickly compute the cross correlation of a structure to a target density map much faster than previous methods. This speedup allows for the analysis of very large structures (millions of atoms) and very long (microsecond range) trajectories, and at a much finer level of detail than was previously feasible. The Timeline plugin uses this new fast cross correlation method to visually present the results of the analysis, making it easier than ever to determine the quality of fit for a MDFF simulation and to quickly identify poorly fitting regions of a structure that require further simulation. More information about other latest features of VMD version 1.9.2 can be found on the release page.
VMD 1.9.2 contains several updates for the Molecular Dynamics Flexible Fitting (MDFF) Method. The mdff plugin contains new options for setting up MDFF simulations with implicit solvent and the new MDFF method for low-resolution x-ray crystallography (xMDFF, recently detailed in this article). Complete details for setting up xMDFF simulations with this plugin can be found in the MDFF tutorial. Additionally, a new graphical user interface for the mdff plugin is now available in the Modeling section of the Extensions menu. This interface allows for easier setup of MDFF and xMDFF simulations, as well as running, connecting to, and analyzing interactive MD simulations. VMD 1.9.2 also contains new multi-core CPU and GPU-accelerated analysis capabilities for MDFF, as described in a recent article. The mdffi cc command can be used to quickly compute the cross correlation of a structure to a target density map much faster than previous methods. This speedup allows for the analysis of very large structures (millions of atoms) and very long (microsecond range) trajectories, and at a much finer level of detail than was previously feasible. The Timeline plugin uses this new fast cross correlation method to visually present the results of the analysis, making it easier than ever to determine the quality of fit for a MDFF simulation and to quickly identify poorly fitting regions of a structure that require further simulation. More information about other latest features of VMD version 1.9.2 can be found on the release page.