Ribosome
The translation of genetic information into proteins is essential for life. At the core of this process lies the ribosome, a quintessential large (2.5-4.5 MDa) molecular machine responsible for translating genetic material into functional proteins. In a growing cell, ribosomes comprise up to half of the net dry weight. Because of its fundamental role in the cell, 50% of all efforts to develop antibiotics target bacterial ribosomes, taking advantage of the structural differences between bacterial and human ribosomes. The Theoretical and Computational Biophysics Group (TCBG) has a long-standing history of investigating the structure and function of ribosome. In collaboration with leading experimental scientists, TCBG researchers have also studied several ancillary proteins, which through their interplay with ribosome facilitate the process of protein synthesis and protein installment in the case of membrane proteins. Elongation factors EF-Tu and EF-G as well as membrane protein insertases such as SecYEG and YidC are examples of these studied ribosomal ancillary proteins.
Spotlight: Antibiotic Resistance (Dec 2015)
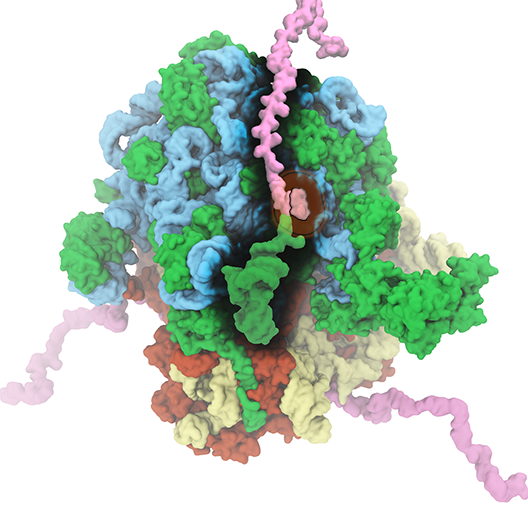
image size:
1.1MB
made with VMD
The ribosome is the ubiquitous machine in all living cells responsible for translating the cell's genes into functional proteins. The majority of antibiotic drugs target the ribosomes of bacterial cells while leaving human ribosomes unharmed. An example are the most widely-prescribed antibiotics, erythromycin and telithromycin. They kill bacteria by changing the properties of bacterial ribosomes and, thereby, disturb the bacterial protein production (see the Oct 2014 highlight Antibiotic Action on the Ribosome). However, modern bacteria fight antibiotic drugs; exposing them to a specific kind of antibiotic drug for too long will trigger the expression of drug-resistance genes, which protect the bacteria, eventually making the drug useless. Due to historical overuse of antibiotic drugs, clinic antibiotic drugs have experienced today serious drug-resistance problems. In a joint effort of computational and biomedical investigations, reported recently, molecular dynamics simulations with NAMD and systematic mutation experiments showed that the above antibiotics interact in a bacterial ribosome with a drug resistance gene - coded nascent protein and make it stall translation; however, engineered simple mutations in the bacterial gene can abolish stalling and, thereby, prevent the effect of drug resistance genes. The research suggests that engineered mutations might be a strategy to prevent antibiotic resistance. Read more on our Ribosome website.



