Ribosome Spotlights
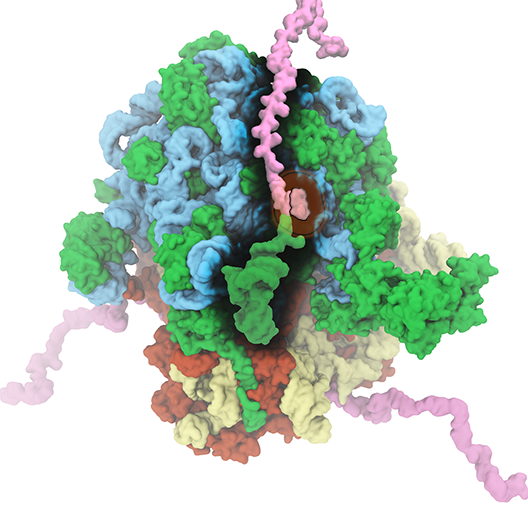
image size:
1.1MB
made with VMD
The ribosome is the ubiquitous machine in all living cells responsible for translating the cell's genes into functional proteins. The majority of antibiotic drugs target the ribosomes of bacterial cells while leaving human ribosomes unharmed. An example are the most widely-prescribed antibiotics, erythromycin and telithromycin. They kill bacteria by changing the properties of bacterial ribosomes and, thereby, disturb the bacterial protein production (see the Oct 2014 highlight Antibiotic Action on the Ribosome). However, modern bacteria fight antibiotic drugs; exposing them to a specific kind of antibiotic drug for too long will trigger the expression of drug-resistance genes, which protect the bacteria, eventually making the drug useless. Due to historical overuse of antibiotic drugs, clinic antibiotic drugs have experienced today serious drug-resistance problems. In a joint effort of computational and biomedical investigations, reported recently, molecular dynamics simulations with NAMD and systematic mutation experiments showed that the above antibiotics interact in a bacterial ribosome with a drug resistance gene - coded nascent protein and make it stall translation; however, engineered simple mutations in the bacterial gene can abolish stalling and, thereby, prevent the effect of drug resistance genes. The research suggests that engineered mutations might be a strategy to prevent antibiotic resistance. Read more on our Ribosome website.
A major activity in living cells is the manufacture of new proteins. For this purpose cells utilize hundreds of ribosomes that read genetic material and according to the genetic sequence synthesize new protein (See also Managing the Protein Assembly Line, Born to Control, Shutting Down the Protein Factory). This crucial synthesis is highly controlled, in particular, in regard to avoidance of errors. For example, proteins of all living systems are made solely of so-called L-amino acids and not the closely related D-amino acids, L- and D-amnio acids being related like left and right hand. Life could have emerged from either "left-handed" or "right-handed" amino acids, but in living cells on Earth, protein synthesis occurs exclusively with L-amino acids, despite of the fact that D-amino acids are actually abundant in organisms and there are neither geometric nor energetic reasons preventing D-amino acid incorporation. For example, D-Serine in the human central nervous systems are present at very high concentrations in vivo. So, the ribosome must have developed early on in evolution mechanisms that prevent incorporation of D-amino acids into nascent proteins. In a recent report, experimental and computational biologists reported their discovery how the ribosome readily discriminates between L- and D-amino acids within its catalytic center. Read more on our Ribosome website.
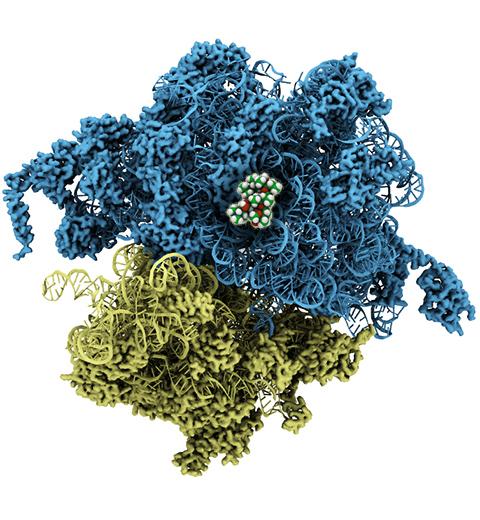
image size:
210.2KB
made with VMD
The ribosome, one of the ubiquitous molecular machines in living cells, is responsible for the critical task of translating the genetic code into functional proteins (See also Managing the Protein Assembly Line). The bacterial ribosome is the target of over 50% antibiotic drugs, for example, the clinically important macrolide family, including the widely-prescribed erythromycin (ERY) which is on the WHO essential medicines list. The antibiotic action of macrolide drugs has been known for over 50 years, however, the molecular mechanisms underlying the effects of these drugs are still unknown. It was previously believed that the antibiotic action by macrolide drugs has to be assisted by the presence of a nascent protein inside the ribosome. However, in a recent study, computational investigations jointly with biochemical experiments have revealed that the macrolide drugs can take an antibiotic action by altering the structure of the bacterial ribosome before translation of nascent protein really begins. Please see more highlights on translational control of the ribosome: Born to Control, Shutting Down the Protein Factory. Read more on our Ribosome website.
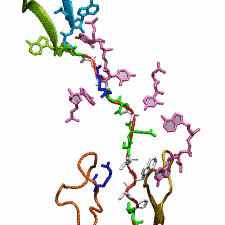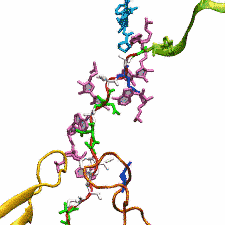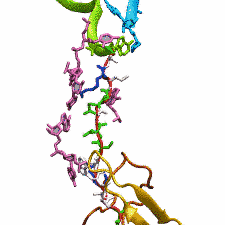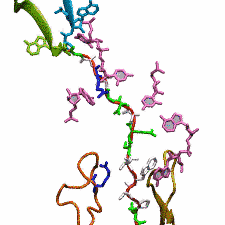
image size:
299.9KB
made with VMD
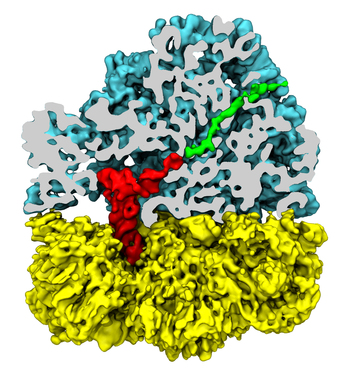
The ribosome functions as a cellular protein factory, synthesizing practically all the proteins in the cell based on blueprints read from DNA (see the April 2012 highlight and Dec. 2009 highlight). However, unlike an assembly line, the ribosome has no foreman directing it. Instead, regulation of protein synthesis is managed by a number of external, and internal, signals. For example, the protein TnaC halts its own synthesis in the ribosome to promote that of another protein (see the May 2010 highlight). Similarly, synthesis of the protein SecA, a translocase that aids in pushing newly made proteins across membranes, is controlled through the nascent protein SecM. Regulation of SecA levels is the only function of SecM, which is degraded as soon as it leaves the ribosome. It is the stalling of one ribosome by SecM that provides enough time for secA, which resides on the same messenger RNA as secM, to be translated by a second ribosome, thus upregulating SecA production. When enough SecA has been produced, it pulls on the portion of SecM outside the ribosome, relieving its stalled state. The critical interactions that cause stalling have now been identified through a combination of molecular dynamics and cryo-electron microscopy via MDFF and NAMD. As recently reported, these interactions form a relay connecting SecM in the exit tunnel to the ribosome's key catalytic center, preventing synthesis and thus explaining how SecM stalls inside "its" ribosome. Additionally, steered MD simulations revealed how SecA can cause the nascent SecM to become unstuck, by breaking those same interactions. More details are provided on our ribosome website.
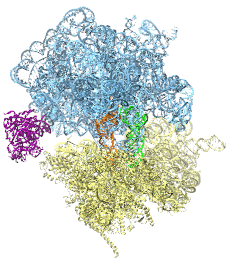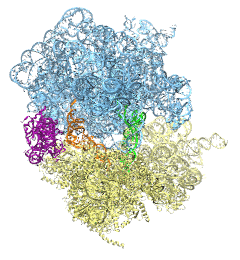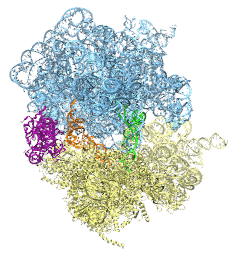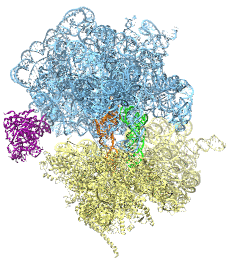
image size:
488.4KB
made with VMD
The ribosome is the protein assembly line in all living cells. The building material for new proteins is supplied by RNA molecules, called tRNA. They enter and move through the ribosome, each adding a new amino acid to the nascent proteins according to the genetic sequence provided through so-called messenger RNA. During the tRNAs' translation through the ribosome, the ribosome itself is not static either. A ratcheting motion and other large scale motions can be observed. However, the exact tRNA and ribosome motion were not clear. Using images from cryo-electron microscopy, MDFF, a computational method based on NAMD, allows one to see the moving parts within the ribosome in great detail. MDFF (see the June 2008 highlight) already provided crucial and unique insights into different aspects of protein synthesis, such as translational arrest of the ribosome by a nascent chain or translocation of an emergent protein across a membrane. In the work reported recently, MDFF revealed the presence of previously unseen intermediate states of the ribosome and its bound tRNAs during the ratcheting motion. A thorough analysis of these states pictures the ribosome as a molecular machine using Brownian motion for its function. More on our ribosome website.
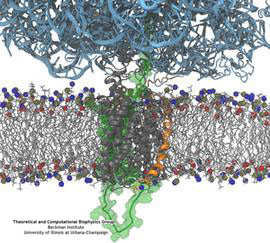
Constructing and correctly placing new proteins is a complicated task for living cells. Starting with nothing more than a sequence of DNA, the cell has to translate the genetic code, stitch together the constituent amino acids, and then place the newly made protein where its function is needed. To accomplish these feats, the cell using tools such as the ribosome, a protein factory (see the Dec. 2009 highlight Managing the Protein Assembly Line), and the protein-conducting channel, a switching station within the membrane (see the Nov. 2008 highlight Patching a Leaky Channel). All instructions for making a nascent protein and localizing it, e.g., to the watery cytoplasm or the oily membrane, are carried within its DNA sequence, but how it is read and then executed has long been unclear. Now, two recent research advances picture these processes in astonishing detail. The first advance (reported here), from a collaboration between cryo-electron microscopists and computational biologists using MDFF, shows an atomic level structure that caught the ribosome in the act of inserting a protein into a membrane. The structure reveals the newly forming protein transiting from within the ribosome to the channel and then through an open gate in the protein-conducting channel into the membrane. The second advance (reported here), accomplished with the simulation program NAMD, explained how the ribosome and the protein-conducting channel manage to pay the energy price of inserting a new protein, one amino acid a time, into the membrane. For more information see our protein translocation website.
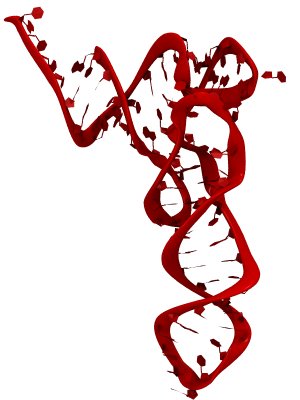
image size:
140.2KB
made with VMD
The ribosome is a molecular machine ubiquitous in all living cells and translates genetic information into proteins. Proteins are made of twenty different amino acids, strung in a linear sequence. The amino acids are coded for by the genes in DNA, but for the purpose of protein synthesis genes are transcribed into a working copy, a messenger RNA. The latter is translated by the ribosome into proteins with the help of transfer RNAs, which bring the individual amino acids. There is a transfer RNA for each of the twenty amino acids. Much progress has been made regarding the static structure of the ribosome, transfer RNA, and nascent protein components (see also the Dec 2009 and Jan 2009 highlights Managing the Protein Assembly Line and Open Sesame). Now researchers are looking into the inner workings of the whole system combining various experiments and computational modeling using NAMD and VMD. The combination yielded unprecedented detailed views of the ribosome in action as reported recently, namely how a dynamic part of the ribosome helps guiding transfer RNAs on their way out of the ribosome, and explains why transfer RNAs behave differently on their journey, depending on if they start synthesis of a protein or if they elongate a protein. More on our ribosome website.
The ribosome is the protein factory of all cells. While proteins are being synthesized, they must travel through a tunnel inside the ribosome before they reach their required location in the cell. This tunnel recognizes certain sequences of nascent proteins and responds to them in various ways. For example, certain protein sequences recognized in the tunnel can shut the ribosome down, stopping protein synthesis. A much studied example is the bacterial protein TnaC, which by shutting down its own synthesis turns on the synthesis of proteins involved in the degradation of a molecule called tryptophan. The structure of TnaC inside the ribosome was previously determined through crystallography, electron microscopy, and computer modeling (see the December 2009 highlight on Managing the Protein Assembly Line). Simulation of TnaC dynamics inside the ribosome using NAMD and VMD has now revealed the mechanism by which TnaC shuts down the ribosome as reported recently. Answered was also the question of how the ribosome recognizes the sequence of TnaC and why many proteins that have sequences similar, but not identical, to that of TnaC do not shut down the ribosome. For more details, see our ribosome website.
Living cells contain millions of proteins composed of sequences of different amino acids that typically fold spontaneously into well-defined three-dimensional conformations and then carry out their role as molecular machines serving manifold functions in the cells (see the protein folding highlight). The synthesis of the proteins is carried out by the ribosome, one of the largest molecular machines present in all cells, which reads the cell's genetic information for the purpose. Three researchers were recently awarded the 2009 Nobel Prize in Chemistry for the determination of the ribosome's structure. The physical mechanism of the ribosome, the cell's protein factory, is still largely unknown. Just as in any factory, there are multiple directions and controls on the protein assembly line. Sometimes the protein products need to be redirected to different parts of the cell, and other times assembly needs to be halted altogether. Now, significant new insights into both of these aspects of protein assembly have been made by combining electron microscopy with molecular dynamics simulations using the recently developed molecular dynamics flexible fitting method (MDFF, see the June 2008 highlight). This combination allowed researchers to visualize the complex between the ribosome and a protein-conducting channel that directs proteins into and across membranes for both a mammalian system (reported here) and a bacterial system (reported here). Amazingly, despite the evolutionary distance between mammals and bacteria, both complexes are remarkably similar. Simulations of the bacterial ribosome-channel complex, among the largest ever performed, further revealed the steps in the direction process. In a third study (reported here), researchers determined how TnaC, as a protein newly synthesized by the ribosome, can stall the ribosome from within during its own assembly, which then controls the expression of related genes. More information on these unique protein assembly controls can be found on our ribosome and our protein-conducting channel websites.
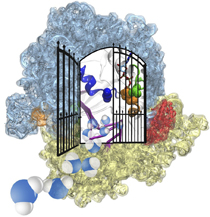
The ribosome is one of the largest molecular machines present in hundreds to thousands of copies in every cell, in charge of synthesizing every protein in the cell faithfully from genetic instruction. For this purpose the ribosome "reads" the sequence of bases on so-called messenger RNA, three bases at a time and depending on the base triple, the codon, elongates a nascent protein by one of 20 possible amino acids, avoiding to an impressive degree adding a wrong amino acid. So far one knew that the reading is done by transfer RNA molecules that have "foots" which match the possible codons and a "head" that brings along the associated amino acid. Each amino acid has its transfer RNA, the transfer RNAs checking if the next codon is "theirs," and if it is they add the proper amino acid to the nascent protein, elongating it. But how does the ribosome make the critical decision at the decoding center, namely if the transfer RNA "foot," the so-called anticodon, matches the codon? The answer is not known, but a key detail has now been discovered through a combination of electron microscopy and molecular dynamics simulation using NAMD, VMD, and a method called flexible fitting (MDFF, see the June 2008 highlight). It was known that a third molecular system is involved, called the elongation factor Tu (EF-Tu), which generates a key signal to the ribosome and transfer RNA through a chemical reaction. This reaction involves chemically attacking a substrate of EF-Tu, the molecule guanosine-triphosphate (GTP), with water, breaking a bond and turning GTP into guanosine-diphosphate (GDP). The puzzle was that EF-Tu is far away from the decoding center. The collaboration between experiment and simulation, reported here, revealed that "correct recognition" through anticodon-codon binding opens a gate in the EF-Tu that allows water access to the GTP inducing the signaling reaction. The finding promises to now establish how the decision at the decoding center is made and how an "open sesame" order is transmitted to EF-Tu. More on our ribosome website.
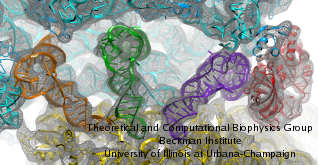
image size:
117.5KB
made with VMD
Living cells are brimming with activity, much of it due to their manifold molecular machines pulling cargo, importing and exporting molecules, digesting food molecules and transforming their energy, reading and copying genetic messages, or synthesizing proteins from these messages (the latter done by the ribosome). Static structures of the molecular machines have been resolved through crystallography: machines pressed into the confinement of crystals and frozen into inactivity reveal their atomic level geometry through this methodology. However, many machines, for example the ribosome, undergo large conformational transitions during their cyclic action, but active motions are hard to view in atomic detail. A way out is offered by electron microscopy which freeze-shocks machines into states characteristic for action cycle intermediates. Unfortunately, the method does not yield atomic resolution images, leaving the chemical detail needed for a comprehension of the mechanisms blurred. Computational methods can be used to bridge the resolution gap: atomic level structures of non-functional states of the machines captured in crystals are deformed to match electron microscopy images. Until recently, the method worked well only for very small machines. Now a team of electron microscopists and computational biologists using NAMD extended the method to common size machines and reported its successful application to the ribosome, providing astonishing detail about ribosome dynamics and function. For more details, see our MDFF website.
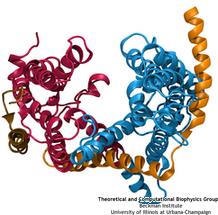
image size:
111.2KB
made with VMD
Everyone knows oil and water don't mix. Proteins observe this rule, too, some choosing to stay in the watery cytoplasm and others choosing the oily membrane. But getting into the membrane is not easy, and most newly formed proteins require another protein, the membrane-bound translocon, to help them insert into the membrane. The translocon, surprisingly also serves as a conduit for proteins across the membrane, thus carrying out a unique dual function. The structure of the translocon showed evidence of a likely "lateral gate", i.e., an exit from the channel into the membrane. How the channel opened to the membrane though, and how it closed afterwards, were not clear from the structure alone. Now, molecular dynamics simulations performed with NAMD, covered in a recent publication, have permitted researchers to understand how the channel opens laterally, how it closes, and how the oily lipids are prevented from invading the water-filled pore. Furthermore, the novel simulation technique, residue-based coarse graining, allowed the researchers to simulate the lipid-channel interactions for up to one microsecond, clearly illustrating that the lipids do not want to mix with the channel interior. More information on these results can be found on the Protein Translocation website.
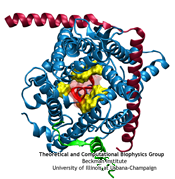
image size:
40.4KB
made with VMD
Anyone who has attempted to fit a long piece of thread through a needle's eye realizes how difficult fitting something so small and flexible into such a small hole can be. Yet this action is carried out every second in every living cell. Flexible polypeptides, proteins, often have to cross a cellular membrane to get to their correct location, whether that location is an organelle within the cell or even outside of it. To accomplish this, they are pushed through a protein pore in an unfolded conformation much like a long string. The channel that accepts the string-like proteins, the protein translocon, allows only certain proteins to pass, while restricting access to molecules even much smaller than the macromolecular proteins. As reported in a recent publication, computer simulations using the molecular dynamics program NAMD helped to answer the question of how such a small channel could achieve this feat, demonstrating how the channel itself can be flexible yet resilient during a protein-crossing event and also elucidating in part how it can maintain such tight control over what is permitted to cross. For more information, see our Protein Translocation website.