Over 1,000 citations of NAMD reference paper
March 29, 2010
The NAMD developers thank our users for 1,000 citations of our 2005 reference paper:
James C. Phillips, Rosemary Braun, Wei Wang, James Gumbart, Emad Tajkhorshid, Elizabeth Villa, Christophe Chipot, Robert D. Skeel, Laxmikant Kale, and Klaus Schulten. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry, 26:1781-1802, 2005.
NAMD is a parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems. NAMD scales to hundreds of processors on high-end parallel platforms, as well as tens of processors on low-cost commodity clusters, and also runs on individual desktop and laptop computers. NAMD works with AMBER and CHARMM potential functions, parameters, and file formats. This article, directed to novices as well as experts, first introduces concepts and methods used in the NAMD program, describing the classical molecular dynamics force field, equations of motion, and integration methods along with the efficient electrostatics evaluation algorithms employed and temperature and pressure controls used. Features for steering the simulation across barriers and for calculating both alchemical and conformational free energy differences are presented. The motivations for and a roadmap to the internal design of NAMD, implemented in C++ and based on Charm++ parallel objects, are outlined. The factors affecting the serial and parallel performance of a simulation are discussed. Finally, typical NAMD use is illustrated with representative applications to a small, a medium, and a large biomolecular system, highlighting particular features of NAMD, for example, the Tcl scripting language. The article also provides a list of the key features of NAMD and discusses the benefits of combining NAMD with the molecular graphics/sequence analysis software VMD and the grid computing/collaboratory software BioCoRE. NAMD is distributed free of charge with source code at www.ks.uiuc.edu.
This paper is available from our website or directly from the journal.
The NAMD License Agreement requires that this paper be cited by any published work which utilizes NAMD. Proper citation is essential to continued NIH funding for NAMD development, as it is a primary way in which we demonstrate the value of our software to the scientific community. As of March 2010, over 1700 publications have cited either the 1999 or 2005 reference papers.
Below are a few of the many biomedically relevant discoveries made by internal and external NAMD users.
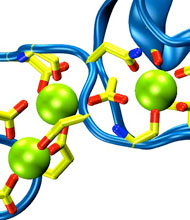
Harvard University and Harvard Medical School News Alert: Our senses are essential for survival and for the exploration of natural environments, and much has been learned about the molecular basis of vision, olfaction, and taste. Yet only a few of the molecules mediating touch and sound perception have been discovered. Now, a team from MCB and HMS has resolved the molecular structure of cadherin-23, a key protein important for sound perception. They have used this structure together with molecular dynamics simulations to understand its mechanics and its function in hearing and deafness.
With the cadherin-23 structure in hand, the team used molecular dynamics simulations to determine its elasticity. The tip link has been assumed to be a relatively elastic, spring-like molecule. However, an extensive set of atomistic simulations, performed using parallel supercomputers with thousands of processors and the NAMD software developed at UIUC, suggest otherwise. The simulations revealed a stiff cadherin-23 molecule, with tightly-bound calcium ions preventing mechanical unfolding. The tip link is therefore suggested to be a stiff cable conveying force to transduction channels, with some undetermined molecular component providing the necessary elasticity for the system.
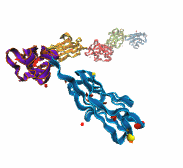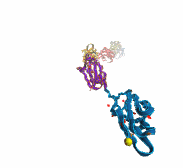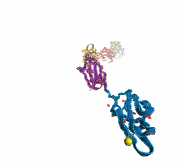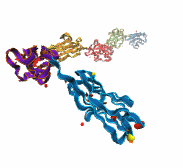
image size:
212.3KB
made with VMD
A smart strategy usually involves a plan B. As it turns out, the muscle proteins in our bodies responsible for the physical motions like running or the beating of our hearts, also rely in their function on having a plan B strategy. When contracting and extending, muscle fibers generate tremendous forces that need to be buffered to protect muscle from damage. This role falls to the muscle protein titin, which is composed of a chain of linked domains, making it a molecular rubber band. When a small force is applied, titin employs its plan A and stretches apart without unraveling its individual domains (like what the movie on the side shows). When a stronger force is applied, plan B kicks in and further elasticity is generated by the unwinding of the protein domains one at a time. By practicing two modes of response to different levels of forces, titin provides the elasticity that muscle needs at a minimal structural cost. A recent computational-theoretical investigation has provided a molecular view on how titin's two plans work, the study featured in a journal cover. The needed simulations were performed using NAMD. Principles described in this study can also be found in other mechanical proteins, recently reviewed here. More on our titin IG6 website.
NCSA News Release: Biophysicists at the University of Pennsylvania used NAMD running on NCSA's Abe to clarify a mysterious interaction between cholesterol and neurotransmitter receptors. Research into how anesthesia works may eventually unlock not only that mystery but dozens of others as well. "Anesthetics have improved significantly over the last hundred years, but the mechanism of anesthesia is not understood at all," says Grace Brannigan, a researcher at the University of Pennsylvania's Center for Molecular Modeling (CMM).
To gain insight into how anesthetics work, a team consisting of Brannigan and fellow researcher Jérôme Hénin, University of Pennsylvania professors Michael Klein and Roderic Eckenhoff, and Richard Law of the Lawrence Livermore National Laboratory, is focusing on the nicotinic acetylcholine receptor (nAChR). This receptor, found in both brain and muscle cells, is a ligand-gated ion channel. The channel opens or closes in response to binding with a chemical messenger (ligand) such as a neurotransmitter, like acetylcholine. When the channel is open, ions can cross the membrane. Anesthetics are believed to close the channel, thus reducing sensations and possibly causing the memory loss associated with being under general anesthetic.
The researchers hope their work leads to improved drug design. "You could fine-tune the properties of the drug if you could understand how the mechanism works," says Brannigan. "For instance, by understanding how anesthetics work, you could design new anesthetics that could be more powerful yet maybe wouldn't have some of the side effects that current ones do."
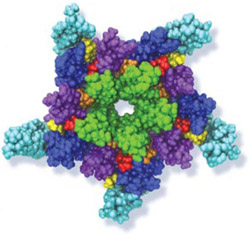
Many living cells, so-called eukaryotic ones, organize their genetic materials in the cell's nucleus, enveloped by a double membrane with guarded access through pores that involve an amazing filter. Like an ordinary filter it permits passage of small particles (biomolecules), but not of large particles (e.g., proteins). However, certain large particles, proteins called transport receptors, can pass. The filter is made of long "finger" proteins anchored inside the pores. The transport receptors can intermittently widen the filter. But to observe how this is achieved is difficult since the finger proteins are highly disordered. As reported recently, simulations using NAMD suggest now a simple and elegant answer: the finger proteins bundle in groups of 2 - 6 and form a brush, filling with its bristles the nuclear pores. The bristles are bundles of finger proteins and have two key properties: (i) on their surface they are dotted with spots of amino acid pairs, phenylalanine and glycine, that are known to interact favorably with transport receptors (see the Aug 2007 highlight, the Feb 2007 highlight, and the Jan 2006 highlight); (ii) the bristles are also interconnected, namely where finger proteins change from one bundle to another bundle, which they do with some frequency. It appears then that the bristles of the nuclear pore filter form an energetically favorable environment for transport receptors. A recent report of new simulations shows that transport receptors are pulled into the bristles of the nuclear pore filter. More information here.
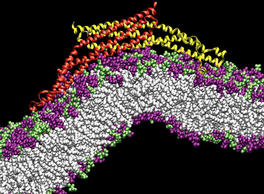
PSC News Release: University of Utah chemist Gregory Voth and grad student Phil Blood are using PSC’s Cray XT3 to tackle a basic question of endocytosis—the life-sustaining process by which cells absorb material from outside the cell by bending their membrane to form a “vesicle” and engulf it. All animal cells depend on endocytosis, which involves various steps, but begins with curvature of the membrane.
BAR domains are a family of banana-shaped proteins shown to bind to cellular membrane as it curves. Experiments suggest that BAR domains mold their concave surface to a section of membrane and induce a corresponding curvature. Voth and Blood undertook molecular dynamics simulations to look more closely. With the XT3 they’ve been able to run efficiently, using software called NAMD, with as many as 1,024 processors. “The XT3 has been amazing,” says Blood. “We haven’t found a hard limit on scaling up the number of processors.”
They used TeraGrid systems at SDSC, NCSA and University of Chicago/Argonne to construct a model and to explore how long a stretch of membrane they needed for curvature to occur. Their final simulations used the XT3 to include the protein with a 50-nanometer length of membrane—probably the longest patch of membrane ever simulated—for a total of 738,000 atoms. Their results, reported in Proceedings of the National Academy of Sciences (2006), show that the orientation of the BAR domain as it attaches to the membrane determines the degree of curvature.
SDSC News Release: SDSC and UC San Diego researchers are using NAMD to zero in on the causes of Parkinson's disease, Alzheimer's disease, rheumatoid arthritis and other diseases. The April 2007 FEBS Journal cover story offers—for the first time—a model for the complex process of aggregation of a protein known as alpha-synuclein, which in turn leads to harmful ring-like or pore-like structures in human membranes, the kind of damage found in Parkinson's and Alzheimer's patients. The researchers also found that the destructive properties of alpha-synuclein can be blocked by beta-synuclein—a finding that could lead to treatments for many debilitating diseases.
Lead author Igor Tsigelny, SDSC researcher and project scientist in chemistry and biochemistry at UCSD, said that the team's research helped confirm what researchers had suspected. “The present study—using molecular modeling and molecular dynamics simulations in combination with biochemical and ultrastructural analysis—shows that alpha-synuclein can lead to the formation of pore-like structures on membranes.” In contrast, he said, “beta-synuclein appears to block the propagation of alpha-synucleins into harmful structures.”
“This is one of the first studies to use supercomputers to model how alpha-synuclein complexes damage the cells, and how that could be blocked,” said Eliezer Masliah, professor of neurosciences and pathology at UC San Diego. “We believe that these ring- or pore-like structures might be deleterious to the cells, and we have a unique opportunity to better understand how alpha-synuclein is involved in the pathogenesis of Parkinson's disease, and how to reverse this process.”