Historical Series: Bionanotechnology
Bionanotechnology and the Computational Microscope
By Lisa Pollack
December 2013See other related articles by L. Pollack
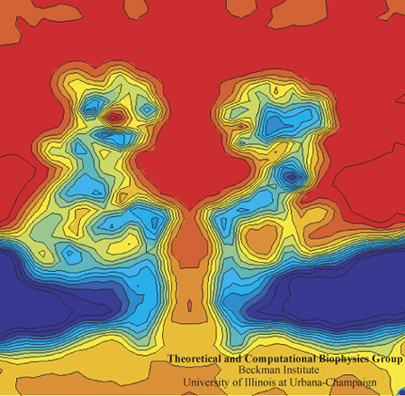
The computational microscope provides a rare view of the electric field inside a nanopore of alpha-hemolysin.
Imagine how the prospectors who descended on California in 1849 would have fared had they possessed a functionalized hand that could simply reach into the Sacramento River and collect its gold. With their pans and sluice boxes the miners still missed the tiniest gold particles–that is, they had no way to capture the nano-sized flecks of the precious metal that graced the many rivers of the Sacramento River Valley. But what if a synthetic protein was crafted that specifically bound to gold and could aid in trapping gold nanoparticles. When Klaus Schulten heard about just such a mechanism, from the very scientist who had tediously labored to find a peptide sequence that affixed to gold, Schulten was intrigued. This eccentric scientist told Schulten he had indeed found a way to isolate gold from the Sacramento River, but wanted Schulten's help in visualizing and explaining why the protein segment itself bound so well to gold. The situation warranted a tool to literally envisage the nano-world of the gold-binding peptide, and luckily Schulten had such a tool in his arsenal: the computational microscope. When Schulten agreed to illuminate the atomic details of the gold-protein system in 2001, he did not realize that this would also launch his entry into the field of bionanotechnology. But he soon discovered that the computational microscope was virtually ordained for bionano-applications.
Both the computational microscope and bionanotechnology are relative newcomers on the timeline of the history of science. Schulten coined the term “computational microscope” around 2005 to describe the imaging technique that can offer what no other traditional microscope can in terms of viewing nano-systems; furthermore, it is composed of unusual constituent materials. Scientific information from chemistry and physics, clever algorithms, powerful computers, and human imagination are but some of the elements comprising this novel microscope. Ideal for capturing behavior of living objects like proteins or ribosomes, the computational microscope, it turns out, can also illuminate what happens when you bring a biomolecule into contact with an inanimate nanomaterial, such as the protein on gold mentioned above. Bionanotechnology is the marriage between two fields that have usually been studied separately: biotechnology and nanotechnology. When experimentalists started to bring “wet” biological materials into contact with traditionally “dry” nanodevices, the combined systems of “wet” plus “dry” were so small that light microscopes did not have the resolving power to see the resulting interactions. What's more, electron microscopy required freezing and thus could not capture progressing behaviors over time or image under natural (i.e. wet) conditions. Enter the computational microscope and Klaus Schulten's research team, the Theoretical and Computational Biophysics group, located in the Beckman Institute for Advanced Science and Technology at the University of Illinois.
“The Beckman Institute is the United Nations of Science,” says Schulten about the place he has called home for twenty-six years. “All of the different sciences count equally; it's not that one of them is better than the other.” The premise of the institute at its very conception was “multidisciplinary.” And Schulten is clear to point out he chose to go to Beckman because he needed to combine science with computer engineering principles to accomplish his goal of simulating biological systems on a grand scale. On top of that, the science he studied was a combination of biology, physics, and chemistry. His move to Beckman, with its accompanying turn to the engineers, however, was often remarked on with derision by colleagues. Yet Schulten still forged onward.
While Schulten embodies a typical scientist working at Beckman, as he entered the field of bionanotechnology at the start of the new millennium, he was well poised to illuminate such technology since the work often required teaming up with engineers or scientists from different fields to improve the sensors they studied. And while many scientists may shun applied science or technology, considering it not pure enough research, Schulten sees science and technology as partners. So it was natural for him to work on projects involving technology, and develop multidisciplinary collaborations, especially with experimentalists. He has been doing this to some degree all his professional life. And although the theoretical side of bionanotechnology–that is, simulating the creative systems the experimentalists imagine–is just starting to become recognized in this burgeoning field, Schulten says the experimentalists are thrilled to work with him and need little convincing of his potential contributions.
While Schulten entered the simulation side of bionanotechnology early, at that point in his career he had been honing the computer simulation of biological processes for almost two decades previously, and had a well-working technology in his group at Beckman. “Some of the questions that were relevant in bionanoengineering were actually pretty straightforward questions where the computer was already pretty good,” remarks Schulten. “And so basically I saw the computer could contribute to this field.” And what the computer offered was an unraveling of the measurement process going on with all the new and unconventional devices that Schulten's collaborators were fabricating. In the laboratory the scientists would measure things, some never before undertaken by anyone, and the computer could help answer questions such as: What does the measurement mean? Where does it come from? And how should I interpret it? For example, why does it take longer for one end of DNA to go through a channel compared to the other end? Why is methylated DNA more mechanically stable? Why is my kinase sensor not working properly? The computational microscope helped provide answers to all of these questions, as well as many more, and in some cases aided optimization of the measurement process.
The technologies the computational microscope are demystifying in this new field of bionanoengineering touch on some of the hottest topics in biology: simplified and inexpensive DNA sequencing, proteins (the kinases) that often go awry in cancer and disease, and epigenetics. Nanodevices have been around for a long time and many forms of them are even found in common household products. Now nanotechnology is being combined with biological systems to make very good devices. Part of that is because the nanoscale is the smallest scale where one can still measure chemistry. As one nanometer is about the width of a typical protein, sensors can go almost to the size of a protein or a molecule and still take instructive measurements. Basically with such small samples, the bionano-device is tailored for single-molecule sensitivity, which is often desired, and makes this such a promising technology. For example, imagine the headaches saved at every level if biopsy of cancer cells only required a tiny amount, because the sensor for it worked at single-molecule specificity. But this tiny expanse of nanodevice plus biomolecule is the perfect size for Schulten's computational microscope. While nanotechnology is now considered “sexy,” as he once put it, Schulten is clear to point out that he has been simulating nanoscale systems for forty years, for that is the scale at which processes happen in the cell. So a device with dimensions of 10nm × 10nm × 10nm is an ideal fit for the computational microscope.
This article will examine many fascinating nanodevices related to biotechnology, and specifically the role of the computational microscope in elucidating the inner workings of these bionano-applications. A theme running throughout is the multidisciplinary aspect of this work, for Schulten has teamed up with researchers from many disparate fields to work on the technologies revealed here. Indeed, as the Beckman mindset stresses work across many disciplines, this story embodies what happens when creative minds from all sorts of subfields come together to tackle a problem. Although the group at Beckman has worked on myriad projects in bionanotechnology since 2001, Schulten revealed he proceeded with no preconceived master plan. Most of his collaborations have happened by chance; topics appeared for consideration and he took them on. While the word “Technology” is part of the title of the Beckman Institute, Klaus Schulten is fully aware of the historical place of technology in science and how it shaped some of the scientific puzzles he had to solve when he entered this field of bionanotechnology. “Yes, you have to be intelligent and work hard,” he says about moving forward in research. “And you also need good opportunities. And opportunities now come very often through technology.” Schulten believes the many new systems and approaches of emerging technologies challenge the scientist with dilemmas to solve. Indeed, combining nanotechnology with biology in simulations has enhanced the range of the computational microscope, for it has uncovered a riveting array of new vistas, many of which we will visit in the coming pages.
We Don't Even Ignore You
“When I went to Beckman people said: Oh, Klaus, where are you going? You are going to the engineers. This will be your intellectual downfall. Don't do it!” The people, in this instance, were other scientists (read: not engineers) who considered their work pure science, and certainly did not value collaborations with engineers or work focused on technology. At the time Schulten made his decision to become part of the Beckman Institute, around 1987, he was a theoretical physicist who studied biology. To take his research to the next level, he realized that he needed computer technology and input from computer engineers. The ridicule from colleagues about embracing computer technology was not new to Schulten, and did not dissuade him from joining Beckman; however, his path to the University of Illinois (home of Beckman) was anything but straightforward.

The homemade supercomputer Klaus Schulten built in 1988.
In the 1980s Klaus Schulten had a productive career as a theoretical physicist at the Technical University of Munich, where he did some of the most highly-cited work in his career. But for him personally, the atmosphere was a bit odd. He started to use computers more and more for his work in theoretical biology, and he paid a price with his colleagues. “Somehow today people accept computing more and more. But in those days,” Schulten says of the 1980s, “people thought literally that anybody who computes was a primitive person.” Any time Schulten used a computing word with his colleagues in the course of a normal conversation, they made jokes and even sexual innuendoes about it, and Schulten felt like a total idiot. Despite the hostile undercurrents, however, that didn't stop Schulten from undertaking a perilous mission: building his own supercomputer on a meager budget.
The plan to build a homemade supercomputer stemmed from exciting work just down the road from Munich, undertaken by friends of Schulten's at the Max Planck Institute for Biochemistry. By 1987 these friends, Hartmut Michel and Johann Deisenhofer, had crystallized and then determined a high-resolution structure of a membrane protein, a task once purported to be impossible. For this feat, they were awarded a Nobel Prize in 1988. Schulten had a front row seat to all the action, and when the structure was determined it was clear that exciting new calculations on a membrane protein could be run for the very first time. And Schulten wanted to be the first to do them. But a big roadblock stood in his way. For this type of calculation, Schulten would need to commandeer a supercomputer at some center, devoted exclusively for a year to his calculation. No center would be willing to accommodate this request.
Right around this time of the newly unveiled structure of the membrane protein, at the university in Munich, two young students in their early twenties assured Schulten he could indeed have his own supercomputer–they would build it themselves with his grant money. While this story is told in full detail elsewhere (click here to read the history of NAMD), suffice it to say that Schulten took them up on their offer and, risking everything, did finally procure a supercomputer all to himself.
With the promise of such exciting and original calculations to be run, and the unconventional, homemade parallel supercomputer under construction, none of Schulten's colleagues in the Munich physics department paid any attention to him. Many had no interest in hearing about his plans for his computer, and most were just plain unaware. To summarize his treatment in Munich, Schulten here likes to point to a Bavarian saying: “I don't even ignore you.” Ignoring means turning a head to look the other way, “but when you don't even ignore it, it's totally not there,” Schulten explains. “You don't make any attempt even to look the other way because you pretend it's just air. It's just nothing.” Despite the indifference of his colleagues, Schulten marched forward with his plans to start using the computer more as a tool in his research, did not think about possibly finding a better environment to do his work, and even turned down a job offer that was unsolicited. But Schulten's rejection of this job offer was not the end of the story.
Intellectual Downfall in a Big Black Hole
Klaus Schulten had a funny feeling about the way the other physicists were acting after he gave his guest lecture in their department. It was 1985 and Schulten was temporarily living in the United States, on sabbatical from Munich. Since the University of Illinois at Urbana-Champaign was one of the few physics departments in the world at that time actually active in biophysics, Schulten agreed to give this invited talk in Urbana. He thus found himself in a small Midwestern college town, surrounded by flat farmland in all directions, which was a stark contrast to the skyline of New York City where he was spending his sabbatical. In between his lecture and the requisite evening dinner with the Illinois physics faculty, Schulten managed to sneak in a call to his wife. “I said to her: They act so strange. I hope they don't offer me a position, because I don't want to go to this god-forlorn place!”

The big black hole Klaus Schulten observed at the Beckman Institute construction site. Ted Brown, Beckman's first director, is in the foreground. Photo courtesy of the Beckman Institute.
Schulten's instincts were correct, however. Some of the faculty had an immediate meeting with their department committee and were ready by dinner to make an offer. Schulten respectfully declined. But the physicists in Urbana were undaunted and suggested Schulten and his wife, also a scientist, spend a term visiting at the university and getting to know the culture of the place, how they did science, before making a final decision. So Schulten moved his family to the Midwest from Munich in 1987 to give the University of Illinois a closer inspection.
Hours from a major city, surrounded by a “corn desert” as it's called in local vernacular, unrelentingly flat, and near no large water bodies to speak of, Schulten was himself surprised to discover that he very much enjoyed his trial period at the University of Illinois. He admits that many Europeans, including himself, considered the Midwest boring. But it wasn't the landscape that swayed this former urban resident. However, the landscape does contain some of the richest farmland on earth, and Schulten found some riches of his own. In fact, many factors coalesced to change his mind, the most important was probably a hole in that rich dirt.
Upon arriving in Illinois in the spring of 1987, the timing was such that Schulten found himself immersed in an exciting new development taking place at the university: the genesis of the new supercomputer center. In 1985, the National Science Foundation (NSF) announced it would devote $200 million in grants to create four centers in academia to bring supercomputer power to civilian researchers. Up to that time, powerful computers were mostly in the hands of U.S. defense department workers, and some industries, such as aircraft and automobile manufacturers or oil companies. But the four centers created in 1985 were specifically for researchers not in the aforementioned fields; Schulten was the kind of target audience the NSF hoped to reach with its new initiative. In Urbana, one of the original four places chosen by the NSF, the National Center for Supercomputing Applications (NCSA) opened for business in January 1986. Schulten, there in spring 1987, says it was a very fresh and exciting time, with so many new hires on staff with impressive backgrounds. They gave him a wonderful place to work, and he had access to fantastic computing facilities, particularly graphics equipment he made liberal use of. “It was definitely one of the JEWELS in my eyes of this place,” reiterates Schulten about the NCSA at the University of Illinois, “that they had this very concerted computational effort that I had been missing in Munich.”
While intrigued by how the university combined computing and science, Schulten also was drawn to the reputation of the physics department. His wife also felt comfortable in Urbana, with many friends in the chemistry department. However, Schulten points to one defining moment that really cemented his decision to move to the Midwest. His colleagues in Urbana told him that if he accepted their job offer, they could provide him with a laboratory in the new institute. When Schulten asked where it was on campus, they told him it was just being built and they would show him the construction site. What Schulten saw was just a hole in the ground, in the rich black glacial moraine of the Illinois soil. “It was really the largest black hole I had ever seen in my life, and it was so big that I realized, this is really a major institute,” explains Schulten. “So basically my decision to come here was because of the black hole that was supposed to become the foundation of the Beckman Institute.”
The Beckman Way
The roots of the Beckman Institute go all the way back to a small town in Illinois just over an hour's drive north of Urbana. There in the city of Cullom, Arnold Beckman was born, and after serving in World War I, got a BS degree in chemical engineering and a year later an MS in physical chemistry in 1922 from the University of Illinois at Urbana-Champaign. Arnold would go on to get a PhD from the California Institute of Technology, where he worked for a while until he founded his own company, Beckman Instruments. Arnold Beckman was an inventor, and two of his most famous inventions were the pH meter and the DU spectrophotometer. While spending most of his time in California running his company, Arnold Beckman kept close ties to his home state, and served in some capacities at the university in Urbana, on boards and cabinets. He and his wife, Mabel, made a donation in 1980 to the university, which established the Beckman Fellowships for junior faculty.
So when some administrators of the University of Illinois in 1983 sat down to brainstorm about possible new projects that would be of such imaginative scope that they could attract the attention of private donors, and put the flagship university on the cutting edge of the research world, it was natural to approach Arnold and Mabel Beckman. But first they had to formulate an ambitious project for consideration. The resulting 1985 proposal was for a “multidisciplinary” initiative, one which would span physical sciences and engineering, as well as life sciences and behavioral sciences. Part of the proposal submitted to the Beckmans highlighted the already-strong interdisciplinary research going on at the University, and the new supercomputer center under development.
The large nature of the gift the Beckmans finally proffered even surprised the people working hardest to attract the donations for the new institute. At $40 million dollars, this was historically the largest single donation to the University of Illinois up to that point in October of 1985, and was additionally the largest gift made to any public university until that time. Ground breaking began in October 1986, and that winter Klaus Schulten viewed the big black hole that would become his future home, the Arnold and Mabel Beckman Institute for Advanced Science and Technology.
“The Beckman Institute was for me,” says Schulten, “a protectorate for interdisciplinary work, and a very beautiful one at that.” He is clear to point out, in retrospect, that he needed the umbrella of this institute to accomplish the goals he had set for himself as a physicist doing theoretical biology. First of all, he wanted to combine science and technology, in this case using the technology of the computer to aid and complement his studies in theoretical biology. To get science and computer technology together, he needed to go to the Beckman Institute, a place where he no longer heard cries of “it will be your intellectual downfall if you go to the engineers.”

The Beckman Way, according to Klaus Schulten, a resident of the Beckman Institute from its inception.
Schulten does not only want to use technology to facilitate his work, he wants to study it as well, research also consonant with the Beckman Institute. In fact, this article covers many promising new biology-related technologies at the nano-scale. These biotechnologies have been demystified by the computational microscope, a tool combining biology, physics, chemistry, and hardware and software, and honed at the Beckman Institute since 1989. A student of history, Schulten is aware of the close relationship between science and technology on the timeline of modern achievements. Not only has much good science come about through close contact with technology, but science can also contribute to emerging technologies. While Schulten appreciates the historical connection between science and technology, he also points to his roots in rural Westphalia, where he grew up surrounded by farmers. “I am much more a person who loves philosophy and mathematics,” explains Schulten, “but who just comes from a background that is much more grounded in reality, in my case literally in agricultural reality, in dirt.” He sees similarities between himself and his boyhood village farmers, who talked about lofty ideas but also had their feet on the ground. Schulten feels that having his feet grounded in the “dirty business of computing” is the way to get things done.
The atmosphere at the Beckman Institute is best encapsulated by Schulten's quote about it, that “character is important, but not what kind of union card you carry.” He joined the institute for two reasons, to be able to conduct interdisciplinary work that truly treats all partners with respect, and to combine science and technology. He calls this “The Beckman Way,” in that science drives technology, and technology drives science. Schulten likes to point out that one instance of the first category–science driving technology–is how technology can pose new questions to scientists. For example, when first introduced, the atomic force microscope, which worked by a tiny tip tapping the surface of a sample to register a two-dimensional image, scientists discovered the contact with the tip could damage so-called soft biological samples. The question scientists then asked was, how could they re-invent this technology so it would not damage their soft samples? And non-contact mode was born.
Schulten is also clear to point out that the Beckman philosophy he coined is a two-way street, in that technology can drive science–with the obvious case that technology can give researchers the means to do their science. One example of this is the electron microscope, which made a whole subset of biological findings possible in the twentieth century. Closer to home, in Schulten's world, the advances over two and a half decades in parallel computing have allowed Schulten to focus on the question that has motivated him for forty years: How do I describe biological organization? He knows this requires studying processes and societies inside a cell, which for Schulten means considering millions of molecules at once. And the technology of the parallel computer with its accompanying algorithms (many designed by his own group) has enabled Schulten to get closer and closer to his goal of explaining biological organization. Additionally, Schulten is using the technology behind the computational microscope to illuminate the bionanoengineering systems discussed below. His years of tweaking the computational microscope paid significant dividends indeed when it came to bionano-systems.
Technology, in fact, was one of the reasons Schulten got funding in 1990 from the National Institutes of Health, for his NIH Center for Macromolecular Modeling and Bioinformatics, which coincided more or less with his move to the Beckman Institute. These centers are funded by the NIH to create unique, cutting-edge technologies and software to advance the biomedical field. Schulten cites two reasons for his successful grant application. First and foremost was the parallel supercomputer he risked everything to assemble. “I had built this computer and thereby demonstrated that parallel computing can be useful in biology when you model macromolecules,” Schulten offers. Until that time, people were using only single processor computers, and parallel computing was in its infancy. The NIH, according to Schulten, had several centers already working on the premise of going parallel, but Schulten was farthest ahead with his newly constructed and whirring parallel machine. NIH was interested in giving this technology more opportunities to blossom. In fact, without containing his laughter, Schulten likes to point to the site visit for the 1990 grant, where he prominently displayed the homemade computer to show the visitors, and hoped they would be impressed by its blinking light, a sign that it was working hard at its task. He reiterated to the site visitors that he couldn't turn it off, that it had to run for two years continuously. With the abundance of impressive graphics displays available in biomolecular modeling these days, a blinking machine would not be considered of much merit during a site visit. Schulten also thinks his work at the time on the brain was an attractive topic for the NIH. “I think they liked that too,” emphasizes Schulten, “that we were not just a one-trick pony show.” Schulten's center was so successful that he would receive funding every five years for the next twenty-five years.
The Computational Microscope: A Tool for Discovery
“One time I published a paper, and I got a referee report back, from a very famous biologist at Brandeis University, and he wrote: This is the best psychoanalysis of a molecule that I ever read!” Schulten likes to joke about this incident that happened to him, but it really speaks to the power of the computational microscope. He uses the computer as a tool, a microscopy even, to intensely inspect a biomolecule to learn its properties. While the computational microscope may provide a gateway to the many facets of proteins and viruses and ribosomes, which are all active research topics in Schulten's group, incidentally, it is not just a technology for visualizing; it is also a tool for discovery.
It has literally been forty years in the making, the computational microscope. The process that led to photography was also created by a physicist, Louis Daguerre, who labored for many decades before he developed the first daguerreotype in 1839. But unlike the still pictures captured in photography, the computational microscope yields results like moving pictures. It takes snapshots of how the system in question behaves over time, and the subsequent “trajectory” is basically a moving animation of the biomolecules. But what goes into generating such a trajectory? The first stage is information from chemistry, physics, and mathematics. Consider a protein with many atoms. Chemistry provides the forces the atoms exert on each other, for example, from chemical bonds. And then Newtonian physics yields the specifics about how the forces from above make all the atoms in the protein move. Next, mathematics reveals how to compute the interactions and the motions. Couple that with continual refinement of algorithms, and the perpetual increase in computer power over the years, and the computational microscope was honed to the point that it is now in the business of discovery.
In Schulten's group, there are two software products, which are both over twenty years old, that specifically undergird the computational microscope. One is NAMD, a computer program that is tailored to work optimally on parallel machines and is mostly geared towards molecular dynamics calculations. The homemade supercomputer mentioned above was the impetus that propelled Schulten into the world of parallel computing and the creation of NAMD, a history that is told in more detail here. While a light microscope is made of brass tubes and glass lenses, in the computational microscope the brass tubes might be considered NAMD and the glass lenses would be VMD, the other major software product produced in Schulten's group. VMD, or Visual Molecular Dynamics, is a visualization and analysis software and is often used in tandem with NAMD. This graphics software has many tools available to help evaluate molecular dynamics trajectories, and grew out of Schulten's need to visualize and “psychoanalyze” the data he was generating. A comprehensive history of VMD can be found here, which highlights many of its unique features.
Ankyrin, whose mechanical properties were first unveiled in 2005 by Klaus Schulten's group.
To reach the point of categorizing the computational microscope as a tool for discovery, the many factors that define it had to be continually improved, such as simulation time and the size of the system in question. While molecular dynamics can be traced to the 1950s, where scientists first used the methodology to simulate liquids, a big step came in 1977 with the publication of a paper by Martin Karplus, applying molecular dynamics for the first time to a protein. This had a big effect on Schulten. In graduate school at Harvard, Schulten sat down the hall from the two students who also worked with Schulten's advisor, Karplus, on this initial foray into molecular dynamics. “I realized that this computational approach,” Schulten says of the 1977 molecular dynamics paper, “opened new doors to describe problems that you couldn't do with a purely theoretical approach that I had taken until that time.” As mentioned previously, Schulten really paid the price for turning to computing in the 1980s, as he often was made to feel like an idiot by his colleagues. But his relentless efforts, especially at the point at which he joined the Beckman Institute, to get computer engineers involved in his research efforts was one of the reasons the computational microscope grew to the point where he believes it is now a tool for discovery.
Not everyone is convinced, however. Skeptics abound. Even with the 2013 award of the Nobel Prize in chemistry to Michael Levitt, Arieh Warshel, and Schulten's advisor, Martin Karplus, for instigating the use of computer methods to elucidate large complex chemical structures, which is a big validation for molecular dynamics, Schulten is sure there will still be skeptics; but because of the Swedish award, Schulten thinks it may not be as difficult as before. He once wrote a paper to the journal Structure, about the discoveries made via the computational microscope, only to have it rejected. On most of his other papers to this journal he teamed up with experimentalists, but not in this specific manuscript. Schulten objected, and it was clear that there was some disagreement on the editorial board about Schulten's paper, and that some of the board were in support of Schulten. They then reached a compromise, and Schulten was told he could write an “essay” for the journal about this topic. The resulting piece, “Discovery Through the Computational Microscope,” was published in 2009, and is quite fascinating reading.
The main point of the 2009 essay is that molecular dynamics is now sophisticated enough to make discoveries, which are then later verified by experiment; it is not necessarily the other way around anymore. Schulten cites as one of his biggest successes his work on ankyrin repeats. These protein pieces are related to how cells sense mechanical cues. Some mechanosensors in living cells have ankyrin repeats attached to them at one end. When the structure of these ankyrin proteins was solved, it was clear that these molecules had to be mechanically sensitive, and the next obvious question was to determine just how mechanically sensitive they were. This was around 2004. Schulten and graduate student Marcos Sotomayor got immediately on the issue and ran simulations to determine the spring-like behavior of the repeats. They found that the protein was a much more sensitive spring than anything seen before. If this spring were hanging from the ceiling, and a feather was attached to it, the feather would pull the spring right to the floor. And they discovered this first, beating out two other experimental teams the theorists knew were also studying the mechanical properties of ankyrin. While Schulten and Sotomayor were able to publish their spring constant in Structure, ironically the experimental team that confirmed this very sensitive behavior of the repeat got a paper in Nature in 2006, although they had to cite the simulation results from the team in Illinois. This is just one example of many illustrating that discoveries can be made with the computational microscope.
“There are many skeptics, and then there are also many people who are positive,” notes Schulten about molecular dynamics. “And very often, they are the same people.” But how could this be? In a molecular dynamics calculation, forces are always being calculated; the particles of the biomolecule are constantly moving closer together or farther apart and forces are calculated over and over. The hitch is that the forces are described in a very primitive way in order to calculate fast enough. In other words, force quality is a slave to speed.
“If people never did molecular dynamics,” reflects Schulten with all sincerity, “and you would tell them today: This is what we want to do. You wouldn't find a single person in the whole world who would say: This is a good idea. Do it!” That being said, when you are dealing with thousands of atoms, and sometimes millions of atoms, the miniscule detail is not that important, and it is still possible to get critical features right. Schulten also likens it to the attitudes in physics, in which discipline Schulten is trained. “The physicists have been extremely successful with theory, even though they are more qualitative,” he clarifies. He points to two cases, the conductivity of copper, and superconductivity, and the subsequent theories that describe them. They work remarkably well, and accuracy is not such a critical point–same thing with molecular dynamics. “So that is why there's a critique, because it's not a very precise science in a way,” Schulten says about the tension over molecular dynamics.
But the other side to this argument is that there are many things described well by molecular dynamics. Often there is no other way to look at the physical mechanism and all its details. The trajectory, or movie of, for example, a protein, is very often the only way to see how the protein works. And since light microscopes can't see individual proteins, and electron microscopes often freeze their samples, molecular dynamics is the only way to see what is going on at times. “And now the situation today is that people have changed dramatically, to actually being glad that they see the movie,” concludes Schulten about former skeptics. He's even seen instances where manuscripts by experimental groups have been rejected because they didn't include a molecular dynamics simulation. As we will see in the following pages, molecular dynamics sheds light on the physical mechanisms underlying the nano-sensors, and often provides very detailed movies of the inner workings of novel devices in the growing field of bionanoengineering.
Gold Binding Peptides
Klaus Schulten's curiosity was piqued when he finally met Mehmet Sarikaya, most likely during 2001. A professor in the Materials Science and Engineering department at the University of Washington, Sarikaya had a reputation as quite a character, according to Schulten. Sarikaya was very interested in biomemetics, the science of using the principles behind nature's biological systems to solve problems in engineering. What Schulten heard that day was that Sarikaya had isolated a protein segment, or peptide, from a large array of mutated peptides, that actually bound to gold crystals. But his peptide only bound to a certain crystal face of the gold, and Sarikaya didn't know why. In the course of their discussion, Sarikaya phrased the problem thus: We want to purify gold particles in the Sacramento River, but first we need to detect them, and this gold binding peptide I've discovered offers just that tool, but I don't know why it works.
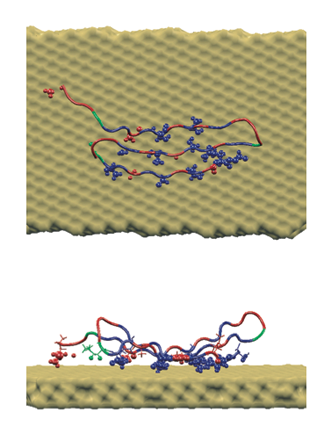
A peptide sequence specifically crafted to bind to gold, which Rosemary Braun and Klaus Schulten illuminated with molecular dynamics.
Schulten realized it would be possible, in theory, to run a molecular dynamics simulation with peptide plus gold, but knew it had never been done before. This would mark his first foray into the world of bionanoengineering, for it combined biology (protein in solution) with a nanosystem (gold particles). And this would begin a long string of collaborations with engineers who designed innovative bio-sensors but had no microscopy available to view the particulars going on at the nanoscale. Schulten realized that molecular dynamics, with the new twist of adding gold nanosurfaces, could help scientists visualize why peptides and gold in solution were forming large flat crystals (that were always the {111} crystal plane). And he had the perfect woman in mind for the job.
Rosemary Braun never anticipated that she would become so fascinated by biology. A physics graduate student at Urbana-Champaign, she only knew she wanted to gravitate to theoretical and computational studies. But Klaus Schulten's class on Non-Equilibrium Statistical Mechanics really resonated for her. “It was FAR and AWAY my favorite class in my entire graduate school experience at Urbana,” Braun remarks. “It was just a phenomenal class.” While the subject matter was in itself interesting, Schulten used examples from biological physics to illustrate the various processes he taught about. Braun was then drawn to biology, and so it seemed obvious to choose Schulten as her advisor when that time came.
Schulten presented to her the problem of the gold-binding peptide, which was the second project she worked on after joining the group. She knew she had to run a molecular dynamics simulation of the entire system, using different crystal faces of gold to find out why the peptide preferred only one specific crystal plane, but there were challenges. The peptide sequence that Sarikaya had isolated was just a mutation, not a well-known peptide whose structure had been determined and studied. To run a molecular dynamics simulation, a structure was necessary; all Schulten and Braun had was the protein's sequence (that is, the order of the amino acids that form the protein segment). Since it wasn't a modification of a naturally occurring protein either, there was no structure like it to even use as a starting point, as a homolog. So Braun and Schulten had to use structure-prediction tools. It should be noted that predicting structure merely from a protein's sequence was then, and still is now, a very difficult process.
Klaus Schulten runs his group in Beckman in such a way that mentoring is an important aspect. Junior people know that they can look to the more senior members for practical, day-to-day advice. Schulten will often ask senior graduate students or postdocs to help undergraduates there for a summer. And the group is full of computer engineers who are actually designing the software (like NAMD and VMD) that most of the members use to complete their calculations, and the developers are a great resource for software questions. Braun was grateful for the extensive guidance she received from Dorina Kosztin, a PhD student and later a postdoc at Beckman. “What he does very well,” says Braun of Schulten, “is facilitate a system where the people who have learned it can teach it in the nitty-gritty way to those who are just starting to learn it. We functioned as a family of sorts.”
In their simulations, Braun and Schulten found out that there was increased affinity for just one crystal plane, and not for other crystal planes. And the computational microscope revealed why: it had to do with the shape of the gold surface. When the peptide was placed flush on one of the other, non-preferred crystal faces, water started seeping in and impacting the interaction between the peptide and the gold surface. But they did not see water get between the peptide and the gold surface when they studied the preferred crystal face, the {111}. Hence, Braun and Schulten could present a detailed description of what Sarikaya was observing, and explain the underlying mechanism with the computational microscope. Little did Schulten realize that this project would become one of many in his new life as an investigator of bionanoengineering phenomena.
Nanopores for Sequencing DNA
“He was like a spiritual bulldog almost,” Klaus Schulten says of the man, Greg Timp, who really pushed Schulten full bore into the field of bionanotechnology. “Every week he wrote an inspirational email, usually with some story from literature, history, the Bible, and then he related it to the work going on here, in nanodevices.” While Schulten was inspired by his colleague Timp at Urbana to contribute to the new field of bionano-devices, there were a few other factors at work that convinced Schulten it would be a good idea to take his theoretical and computational methods into a realm they were not necessarily tailored for. That realm would be the combination of the “wet” biological materials with the “dry” nanotechnology. While Schulten had spent his whole professional life on the former, he was just about to delve into the latter in his upcoming research.

Alek Aksimentiev, a postdoc in Klaus Schulten's group from 2001-2005, who worked on DNA in nanopores.
Part of Schulten's taking the plunge into the nano plus bio admixture stems from his realization of an historical moment in science. While many consider the twentieth century to be the era of physics, now many prominent scientists are calling the twenty-first century the era of biology. “And this era,” Schulten explains, “like any major scientific era, is very often determined by not only pure science developments but also by pure technology developments.” For example, in the twentieth century, one only has to look at the transistor, which flowed from solid-state physics, and led to the computer revolution and all its accompanying technologies. Schulten sees the relatively new field of bioengineering as part of this Life Sciences Century, and one instance where pure science and technology go hand in hand. He saw for himself why technology needed to be combined with the life sciences. “The reason why it's really necessary,” Schulten says of bioengineering, “is that one needs to measure very small samples. And these electrical devices could be scaled smaller, smaller, smaller, to exactly the scale you need in biology.” As living systems are made of molecules, often in low concentrations as there are very few of the same ones in a whole cell, measurement needs to be extremely sensitive, routinely down to the single-molecule level. For this, new technologies need to be employed. And one of the ways to be extremely sensitive and act with very small probes is to utilize nanotechnologies. So not only did Schulten recognize the historical significance of the blossoming field of bionanoengineering, but he knew that the systems (biomolecule plus nanodevice) were so small he could in fact very well simulate the entire thing. He has always lived in his own “nano-village” as he once called it, simulating biological systems considered nano-sized, and these bionano-devices fit nicely into his village rubric.
Another factor at work in bringing Schulten into the field of bionanoengineering is that the University of Illinois at Urbana-Champaign houses the Micro and Nanotechnology Laboratory, and so is a strong presence in the field of bionanotechnology. While the Micro and Nanotechnology Laboratory does photonics and microelectronics, it also focuses on nanotechnology and biotechnology. Schulten was essentially steps away from this hub of cutting-edge research, and thus near experimentalists who were eager to collaborate with him. They could use his expertise to visualize their novel systems, and his work with these researchers from this laboratory will be covered below in detail.
Hence, for Schulten, the time was right, historically speaking, and his presence at Illinois also brought him into contact with experimentalists whose novel devices he could simulate. The first instance of Champaign-Urbana offering Schulten an experimentalist to work with was his collaboration with electrical engineer Greg Timp, the spiritual bulldog. When he started up this collaboration, in 2003, Schulten also had an excellent postdoc who was critical to the success of the mission to combine “wet” biological molecules with “dry” nanodevices in simulation. That postdoc was Aleksei Aksimentiev, and the system he and Schulten worked on from Greg Timp's lab was DNA in a nanopore.
DNA is a fundamental molecule of life and it contains all the information to make a complete living entity, such as a human, plant, or bacterium. Thus DNA is considered hereditary material and is basically the storage information system for living organisms, much as a hard disk or a tape drive stores all the information for a computer. There are basically four letters of the DNA alphabet, and the sequence of those letters codes for genes. So the point of DNA sequencing is to unravel the order of the letters that code for each gene of a living entity. (The sum total of all the genes of a living entity is called its genome.) The ability to determine the sequence of a living system's genes has had huge implications in the fields of biology and medicine. Some of the areas most revolutionized by DNA sequencing are plant sciences, forensic biology, pharmacology, and genetic disease studies.
But DNA sequencing is not cheap, straightforward, or quick. However, great technological strides have been made, and the cost of sequencing an entire genome has gone from $95 million in 2001 to roughly $10,000 in 2011. Many people are working on alternative methods to determine the genetic code. One such method is to use a nanopore.
A nanopore is a tiny pore in a membrane; the diameter of the pore can go from half a nanometer to a few nanometers, and the length of the pore might vary from two to thirty nanometers. A nanopore can be made by nature, such as the channel created across a membrane in a cell, or manufactured in a laboratory, such as the pores Greg Timp made. In Timp's case, he drilled holes in a silicon nitride membrane with an electron beam, and perfected methods to make solid-state pores of various sizes.
The way nanopore sequencing works is the following: Immerse the nanopore in an electrolyte (which is just salty water, or water filled with ions) and apply a voltage difference. To get some idea of scale, if the nanopore were your hand and you were sitting on the 30th floor of a skyscraper, the electrodes would each be the size of a house, and a negative electrode would be on the 50th floor and a positive electrode on the 10th floor. The ions in the solution would begin to flow through the pore and create a steady state current that could be measured. Now if a strand of DNA were introduced into the system, an interesting thing happens. The DNA strand would find its way to the pore and then get sucked in and go through it. It is not necessary to actually bring the DNA to the pore. The DNA would partially block the current as it was going through the pore, and the crux is that the blockade current would be sensitive to each of the letters in the strand. Hence, by looking at the current profile, it would be possible to determine the sequence of the DNA strand. “So in this sense, this nanopore sequencing is analogous to a tape recorder,” says Aksimentiev. “You have a sequence written in the tape and you push it through the reader.”
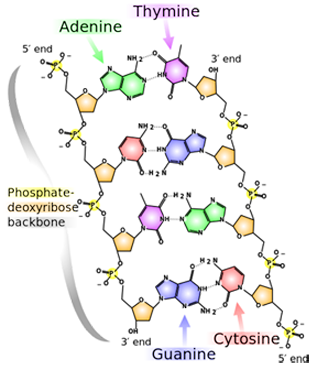
A snippet of DNA, whose four bases are A, T, G, and C. Note the slight structural difference between the 3' end and the 5' end.
One of the most popular methods of DNA sequencing is called Sanger Sequencing. It takes help from biological enzymes that synthesize a complimentary strand to the one in question. “We are relying on biology to read the DNA sequence for us,” Aksimentiev notes. But nanopore technology would no longer need enzymes to manipulate the chemistry of the molecule. However, there are major hurdles with nanopore sequencing. The DNA code is written in atoms, as it is almost impossible to come up with anything smaller than atoms for storing a blueprint of a life form. Shown is an image of a bit of DNA with its four bases (adenine, thymine, guanine, cytosine, or A, T, G, C). The atoms that make up the bases are just carbons, nitrogens, oxygens, and hydrogens. Nanopore sequencing would have to distinguish between nucleotides, or bases, that just differ by a few atoms. “And that, if you think about it, is a really tall order,” says Aksimentiev, “because you are looking at really small differences, atomic scale differences, between very small objects, individual objects. Because we want to read out that sequence of DNA with single nucleotide precision, so for each nucleotide we want to have resolution.”
In 2003, Greg Timp was pursuing the idea of using solid-state nanopores to sequence DNA and wanted Schulten and Aksimentiev to visualize what he was measuring. Since the diameter of the pore was just a few nanometers wide, and the system immersed in liquid, neither an optical microscope nor an electron microscope could do the job of proper visualization of the DNA in motion through the pore. Schulten realized molecular dynamics could probably be used to describe the three-part system of electrolyte, DNA, and nanopore. To Schulten's knowledge, at that time in 2003 no other group had done similar simulations on these kinds of bioengineering platforms.
Today it perhaps seems sensible to get molecular dynamics into the bionano-game, but Schulten took a chance back then. “We could have fallen flat on our faces,” Schulten reiterates. “We had to learn a lot to do things right, gain more expertise, be patient and wait actually for several years until it caught on and people said: Oh, this is interesting.” Schulten explains that when characterizing nature computationally, artificial descriptions are often used, so when a totally new substance is added, things can easily go wrong. With a system of three components, if description of one component is significantly better than the other two, things get out of balance. Thus he and Aksimentiev had to be very careful with their novel system so that each component was described equally well.
And they had some great successes in elucidating the translocation of DNA through the nanopores. One such success came from asking the simple question: What is the smallest pore DNA can snake through? It seems unlikely that a double-stranded DNA piece could fit through a pore whose diameter was smaller than the width of the double helix, but molecular dynamics predicted that it could. The reason is that the DNA gets stretched by the electric field in the very center of the pore. The behavior of the potential inside a pore is only something imaged with the computational microscope; the potential stays the same from one electrode to almost the middle of the pore, and then in the very middle it falls precipitously, and then outside the middle to the other electrode it resumes almost constant value. This behavior right at the center of the pore stretches DNA to almost twice its equilibrium length, decreasing its diameter so that translocation is possible. Molecular dynamics predicted critical pore radii and associated electric fields, which were all verified by experiment. This work that Schulten and Aksimentiev conducted with silicon nitride nanopores was not the only nanopore sequencing work they found themselves immersed in.
Imaging Alpha-Hemolysin: Precursor to Computational Microscope
At the same time Schulten and Aksimentiev were working with solid-state nanopores, they were conducting additional work on another membrane channel–this pore engineered by nature–which led to a very highly-cited paper, a publication Schulten claims is one of the best to ever come out of his group. And this 2005 article really compelled Schulten to later coin the simulations done by his group “the computational microscope.” But it all started with the word “imaging.”
The idea to use a nanopore to sequence DNA or RNA goes back to at least 1996, when a group of scientists (spearheaded by David Deamer and Daniel Branton) published a paper suggesting this novel use of a membrane channel, although the idea may even have percolated for years before the publication. The membrane channel they used was one found in nature, called alpha-hemolysin. Secreted by the bacterium Staphylococcus aureus are building blocks of alpha-hemolysin, which then form a channel in the membrane of a target cell. What results is the uncontrolled permeation of water, ions, and small organic molecules. The targeted cell also can lose its important molecules, as well as its steady potential gradient, and hence this eventually leads to cell death. But alpha-hemolysin, at the turn of the millennium, was recognized as a beautifully engineered transmembrane channel built by nature, so scientists started studying its properties, and then a subset of researchers actually started studying this channel as a potential candidate for DNA sequencing.
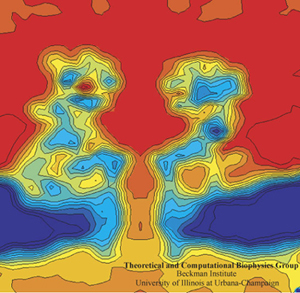
Alek Aksimentiev and Klaus Schulten designed a protocol to view the electric field inside a pore, an image only possible with the computational microscope.
Schulten and Aksimentiev wanted to characterize the properties of alpha-hemolysin embedded in a lipid membrane using molecular dynamics at this time, since its structure had been published and it was a hot topic in biology. But they conceived a clever twist: run a lengthy simulation and look at the average properties of the channel. “That was also much more reminiscent of what people know about the system,” says Schulten about their averaging, “than the constantly fluctuating alpha-hemolysin that you see in a molecular dynamics simulation. It's a very nervous kind of protein, with all kinds of tempers and conformational changes and so on. But on the average it behaves pretty reasonably.”
One of the most important properties they averaged was the electrostatic potential inside the channel. In every living cell there is a negative potential inside, and this potential is critical, as it drives many cellular processes. In the alpha-hemolysin channel all sorts of things flow across it, such as water, ions, DNA, and proteins. When experimentalists were studying alpha-hemolysin they would determine electric currents in the channel by applying external electric fields across the membrane. Schulten and Aksimentiev could reproduce these kinds of experimental studies (called patch clamp measurements) in their simulations. But then they presented a picture of the electrostatic field in these instances. To do this, they ran a long molecular dynamics simulation, and then averaged over the thermal motion that the channel undergoes, with all of its charges that contribute to the field. And in this way they envisioned the electric field inside the channel.
The innovation of this averaging stems from the fact that a biological cell is, electrostatically speaking, extremely noisy. It has ions that diffuse all the time; it has lipids with fluctuating head groups; it has dipole moments in its proteins. Schulten likens a snapshot of this potential to taking a picture of New York City in a rainstorm–you only see the rain, not so much the buildings. But if you take snapshots for two hours and then average, at the end you see the buildings and not so much the rain. Aksimentiev used the tools available in Schulten's group and some ingenuity of his own to introduce this averaging technique to the world. And it showed a beautiful result of the electrostatic potential–on both sides of the membrane, of the surface of the membrane, and also within the channel of the alpha-hemolysin. The result was a 2005 paper, “Imaging alpha-Hemolysin with Molecular Dynamics.” This paper is extremely highly cited, with 252 citations already and counting. The method Aksimentiev and Schulten brought to the scientific community would not have been possible earlier, as advances in NAMD and computer power aided the group in Beckman in 2005. Schulten says it really highlighted the resolving power of the computational microscope. And to his knowledge, this was the first time he used the word “imaging” in relation to molecular dynamics.
Here's a Test for You, Schulten
Klaus Schulten was certainly surprised to hear that if he wanted to work with experimentalist Amit Meller on alpha-hemolysin, first he had to pass a test. But as Meller explained the project in detail, Schulten grew more and more excited, for Schulten realized this “test” would be a great assessment of his life's work in molecular dynamics.
It was May of 2004 and Schulten was at a conference on nonlinear studies in Santa Fe, New Mexico. Amit Meller, then at the Rowland Institute in Massachusetts, was also at the conference and Schulten heard Meller's talk about DNA translocation experiments through alpha-hemolysin. Late one evening at the conference there was an after-talk and beer session, and Meller and Schulten got to chatting. Schulten revealed that he and Aleksei Aksimentiev had been working on simulations of alpha-hemolysin, and that Schulten would love to collaborate with Meller. Meller said he had been working on an experiment, which had a very clean result, but it presented him with a puzzle. He had been unable to explain this microscopically and had been wondering about the explanation of the conundrum quite a bit lately. The dilemma he faced was based on the fact that DNA is not the same from front to back. One end of it is called the 3' end, and the other is the 5' end (as is shown in the image above of the atomic structure of DNA). Meller and team were watching single-stranded DNA enter the nanopore, and in response they measured the residual current that resulted, which was about 10 to 20 percent of the steady state current. “We figured out a way to perform an experiment where we forced our molecules to enter the pore either on the 3' end or the 5' end,” Meller told Schulten that night. “And when we measured the residual current, we saw that there is a difference, and a substantial difference that we could characterize. It was that one of the ends was blocking the current more than the other.”
Moreover, he told Schulten, when they simply let the DNA molecule find its way to the pore and then escape, they found a substantial difference in the escape time for the 3' versus the 5' end. Meller says the reason that this was both surprising and puzzling was that the strands of DNA in his translocation experiments were just homopolymers, that is, 70 nucleotides of adenine (base A) right next to each other. When you flip it over the strands look exactly the same. “So it was quite mind-boggling to realize that nature knows to distinguish between those subtle differences between the entering 3' and 5' ends,” recollects Meller.
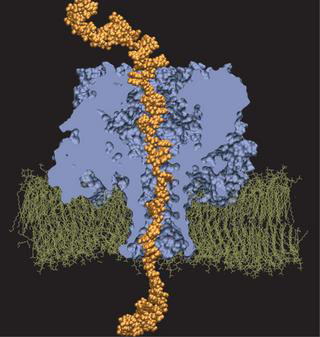
DNA permeation in alpha-hemolysin, as simulated by Alek Aksimentiev and Klaus Schulten.
So Meller clearly laid out to Schulten that there was a very clean and crisp experiment, but that the two distinct ends of DNA behaved differently in both the situations he described. However, he never told Schulten which end did what. There was a reason behind Meller's bold challenge: “But then, maybe I had an extra beer that evening,” he recounts. “I was still a junior faculty at that time, and Klaus was much more, I would say, senior than me.” He told Schulten that while he had a lot of respect for people doing computational experiments, Meller wanted to challenge Schulten. Meller wanted Aksimentiev and Schulten just to run the molecular dynamics simulations and get the results in a completely unbiased way. In other words, the theorists would run their calculations in a double-blind fashion. Meller asked Schulten to perform two tests. First, find out which end blocks the current more. And second, find out which end takes almost twice as much time to escape from the pore. He told Schulten that since there is a 50% chance to get the first question right, and a 50% chance to get the second question right, the overall chance to be correct randomly is just 25%. But if they did the computations correctly, then there would be a potentially good publication as a reward.
Meller did not know Schulten well at that time, and was unsure of how Schulten would react to being challenged like this. But Schulten grew very excited, according to Meller, and seemed to take no offense. And, in true Schulten fashion, even though it was late at night in the western United States and even later in the Midwest, Schulten called Aleksei Aksimentiev in Illinois to tell him to start the simulations immediately. Schulten confesses that he and Aksimentiev thought Meller was a tad crazy, and they thought maybe Meller wanted to trick them. But Aksimentiev did the simulations very carefully and got his answers about three months later.
The scientists in Illinois called Meller on the phone and reported their results. They were correct on both accounts! The 3' end produced the lower residual current, and the 3' end escaped from the pore more quickly than the 5' end. After further discussion, and further ingenious simulations by the Beckman team, the microscopic picture of what was going on was revealed. The answer can be explained by likening the DNA strand to a Christmas tree going through a door. Clearly, there is only one good way to bring that tree through a door. When the strand of DNA was confined in the alpha-hemolysin, the bases all tilted toward the 5' end. Therefore, the friction experienced by the 3' end versus the 5' end was different, and the 3' end acted like the upper tip of the Christmas tree when translocating through the pore. Meller also offers another analog: there is only one good way to pet a cat.
Meller was pleased to have a microscopic explanation of his experiments. And for Schulten and Aksimentiev, who in 2004 had only just embarked on simulations of DNA in nanopores, this was a great validation of the computational microscope, for their careful procedures clearly paid dividends. The team quickly wrote a paper, which also included authors Jéröme Mathé and David Nelson, the former the postdoc of Meller's who did the experiments and the latter who did some theoretical modeling. The result was a publication in the Proceedings of the National Academy of Sciences. This tale, of being challenged to do molecular dynamics in a double-blind manner, and not only getting the correct answers but actually finding the microscopic explanation to the phenomenon, left a big impression on Schulten. As he says, “this was one of the most wonderful stories, actually, in all of my molecular dynamics life.”
Carbon Nanotubes: It's all about Polarization
Graduate student Deyu Lu had a challenging task. With a few studies on membrane proteins and light harvesting under his belt already as a graduate student of Schulten's, he needed to find a project suitable for and worthy of a dissertation. Lu was to present his grand idea to Schulten, and then Schulten said he would give it a grade, A, B, C, etc. Two things simultaneously converged at this time, circa 2002, which earned a grade of A+ from Schulten for Lu's idea.
Unbeknownst to Lu, his assignment of a TA office would have major implications for his future. Right next door to Lu's office sat another physics graduate student, Yan Li. As chance would have it, the two had met once before in China, at a meet-and-greet for students who all were headed abroad to study in Urbana. But with their offices right next door, they really started to get to know each other by doing homework together. Li, while also in the physics department, had chosen to work with electrical engineer Umberto Ravaioli, as her first foray in an experimental physics lab had taught her that she was too clumsy for what was required in that setting. So she realized she needed to find a theorist to work with. Since the theoretical physicist she initially wanted to work with was on sabbatical, she eventually found Ravaioli, who was doing theoretical material science, a field she really wanted to pursue.
Now that Li was settled into a theory group, she began to immerse herself in the literature about carbon nanotubes, the topic her advisor thought would make a great dissertation project. To visualize a carbon nanotube, imagine a flat sheet of honeycomb, and placed at each of the apexes of the hexagons that make up the honeycomb are carbon atoms. Now roll that 2D sheet into a cylinder, and you have the nanotube, whose diameter is anywhere from 1 to 50 nanometers wide. Discovered in 1991, these nanotubes have a host of interesting properties and, a decade later, there were a flood of papers exploring their many fascinating aspects. It was just at this time, when there was much excitement about carbon nanotubes, that Yan Li started investigating them. Her advisor suggested she might study the metal-to-semiconducting transition. Roughly speaking, depending on the structure of the nanotube, it can be either metallic or semiconducting. So Li was just at the beginning stages of her dissertation.

Deyu Lu and Yan Li in 2006, at the UIUC Quad. Photo used with permission from Yan Li.
By this time Deyu Lu and Yan Li had started dating. While Li was pretty sure her dissertation topic would be carbon nanotubes, Lu was still looking for his project idea. Although Lu was by now working in Schulten's group, and as such, doing biophysics-related projects, he had gotten a master's degree in condensed matter physics back in China, and was very interested still in this field. In fact, he often talked to his girlfriend about her carbon nanotube headway. And he grew more and more fascinated by carbon nanotubes. One day he came upon a paper in the journal Nature, reporting that water actually conducts through a nanotube, as was shown in molecular dynamics calculations done by the authors. “I think this is probably the first work that I saw that related carbon nanotubes to some kind of biological environment,” notes Lu. “So I was amazed by the story there.”
Lu started thinking longer and harder about carbon nanotubes at that point. As he had training in condensed matter physics, where proper description of electrons is paramount, he started to think about polarization and its role in the water-nanotube interaction. Polarizability is essentially related to the degree of distortion of an electron cloud when near some kind of charge. Imagining again the honeycomb basis of the nanotube, at each apex the carbon atom is bonded to three other carbon atoms. But carbon has four valence electrons, so at each apex one electron is more or less itinerant. Looking at the whole nanotube now instead of just the individual carbon atoms comprising it, these roaming electrons can form clouds that distort if an electric field is applied; this is one example of how carbon nanotubes can become polarized. But polarizability is also possible if the nanotube were filled with water molecules, which have dipole moments. Lu started wondering about how to give a proper description of the electronic behavior, how to characterize polarizability, and what might be the interaction between water and the nanotube. “So when I started to ask myself those questions, I felt this could be a very promising direction for me to explore,” Lu explains.
During this whole time, Lu was consulting with Yan Li, and they spent considerable time discussing carbon nanotubes, and they bounced ideas off each other. They finally settled on a methodology to use to characterize carbon nanotubes and their polarizability. The next step was to not only propose their idea to their advisors, but also see if they could sell a collaboration between the two groups. This was before Schulten had really immersed himself in the field of bionanotechnology, but Lu reports that Schulten gave him an energetic thumbs-up for the proposed project, and, in the Beckman fashion, was happy to consider a collaboration with Ravaioli. While this collaboration turned out to be very fruitful publication-wise, it also led to other profound realms. Yan Li and Deyu Lu married at the end of 2003.
The overall goal of the study was to look at polarization and then also do some simple molecular dynamics calculations. To understand the polarization, the couple needed to employ an electronic structure method, which, as its name suggests, looks at the electrons of the system to calculate its properties. Electronic structure methods are quantum mechanical, while molecular dynamics is considered classical. Deyu Lu says they settled on the method called tight binding because it was simple enough; one can do lots there with just pen and paper, but it also provides adequate physical insight. While using tight binding may sound straightforward, Schulten is clear to point out that what Lu and Li accomplished was quite a feat, using that level of theory to describe nanotubes. Schulten is quite proud of what was accomplished.
In fact, the couple had to contend with challenges at every level. There was a senior postdoc in Ravaioli's group who did provide lots of advice and help, Slava Rotkin, and Rotkin is even on some papers, but mainly Lu and Li had to figure out everything on their own. Lu says a big source of help to him was the slim book, Physical Properties of Carbon Nanotubes, by Saito, Dresselhaus, and Dresselhaus, which he read cover to cover. Schulten sees the constellation of personalities and groups as a key reason for the success of the nanotube work. “This was one of the really clear cases where it was proven that how well a group does with a project depends very often on the particular personality, giftedness, and ability of an individual,” says Schulten. “And it's not like the group can do anything they want.” Yan Li says she and her husband understood each other very well, and that their similar backgrounds–undergraduate physics majors, same class in graduate school–also played key roles. While the reasons for their success are numerous, it seems clear that the couple brought out the best science in each other.
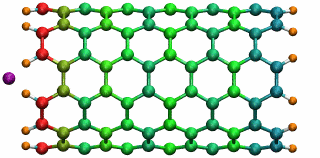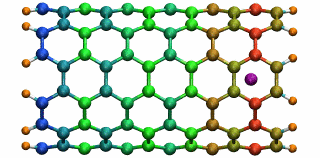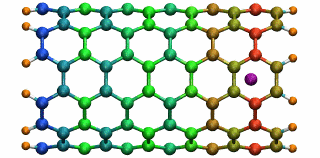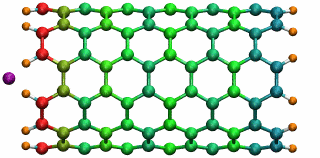
An ion in a carbon nanotube. Once it gets sucked in, the ion oscillates as shown.
Perhaps the highlight of the carbon nanotube study was an exciting observation Lu made, which resulted in a paper in Physical Review Letters, a premiere journal for solid state physics research. In order to see the polarization in a carbon nanotube, a source of external charge needs to be supplied. At the beginning of the work, Lu and Li subjected the nanotube to an electric field, then filled it with water. Deyu Lu started to wonder what else he could do to elucidate the polarizability, for at that point he had written his own simple molecular dynamics code and had examined the water-nanotube system. And then it occurred to him: why not use an ion. He knew that a single ion was a simple enough system to incorporate into his existing molecular dynamics code. He also realized that since water is neutral, he should see an even bigger effect on the polarization if he used an ion.
Deyu Lu can still remember the day he tried his idea out. He put a potassium ion outside the carbon nanotube, and ran his program. The ion was sucked into the tube. Not only that, but then it started to oscillate! The ion couldn't get back out of the tube, a consequence of energy conservation. Lu immediately ran to Schulten's office to tell him. The extremely positive response he got from his advisor was unexpected. Schulten was thrilled. He told Lu that physicists love to see an oscillator, and that Lu and Li should expound on this result and dig deeper. The outcome was a 2005 publication, “Ion-Nanotube Terahertz Oscillator.” Schulten claims it is probably one of the best papers ever to come out of his Beckman group. For Deyu Lu, his project on the carbon nanotubes had lasting implications for his future. Recall, at this point in time, Schulten had not fully immersed himself in nano-engineering projects. Lu feels fortunate that Schulten allowed him to do something slightly different than most of the other research projects in the Theoretical and Computational Biophysics Group at Beckman. “In that regard, Klaus gave me his whole support and just let me do the thing I liked,” says Lu. “And I really feel fortunate for that situation.” But most of all, it fashioned the way he approaches research, and developed his independent thinking, skills that Lu says he relies on to this day.
Nanodiscs–But How Do They Really Look
While Klaus Schulten admits he did not have a structured plan that he followed in order to acquire new projects in nano-engineering, his mere presence as a faculty member at the University of Illinois–a large school–offered him a cornucopia of collaborations in exactly this field. When Schulten was approached by a colleague seeking help in visualizing this biochemist's new brainchild–a type of nanoparticle people thought he was nuts for developing–Schulten realized the computational microscope was a perfect tool for the job. This resulted in one of Schulten's best efforts in bionanoengineering, and is really a concerted University of Illinois effort, from its earliest beginnings to the present.
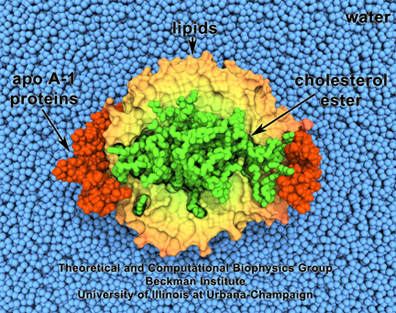
HDL, or good cholesterol. Lipids are surrounded by proteins, and enclose cholesterol.
Steve Sligar, a biochemist at Champaign-Urbana, had acquired an atomic force microscope in the 1990s and little did he know that, in looking for candidates to image with his new machine, it would lead to creation of what he dubbed a “nanodisc.” One of the particles he started studying was a lipoprotein, inspired by pioneering work begun at Urbana by Ana Jonas. Starting in the 1980s, Jonas had been studying the atherosclerotic process and the role of lipoproteins. Most people are aware of lipoproteins as it relates to human health, that good cholesterol, or HDL (high density lipoprotein), circulates in the blood collecting cholesterol in the body. Jonas actually fabricated HDL particles in her lab. She even persuaded Schulten to help her visualize these reconstituted particles in a project they worked on in 1997. HDL look like orbs made of lipid particles, with, for example, cholesterol inside, and a protein on the outside sort of binding the whole thing together, as seen in the image.
Sligar and his team started making their own nanoparticles in the lab, reminiscent of Jonas's HDL particles. Sligar then genetically engineered various proteins (called apolipoproteins, or more instructively, membrane scaffold proteins) to form around the tiny bits of lipids. When the membrane scaffold proteins and the lipids were thrown together, they assembled themselves into structures that Sligar decided to name “nanodiscs.” Then he had a “wild idea,” according to an historical article he penned about nanodiscs, published in 2013. Membrane proteins, as their name would suggest, live in membranes, and studying these proteins has presented immense challenges to researchers. This is because outside of the membrane, the proteins become inactive and die. As mentioned above, in 1988 a Nobel Prize was awarded for actually isolating a membrane protein on its own and then forming crystals of it suitable for structure determination. The Stockholm award speaks to the challenges of working with membrane proteins. At this point, around the dawn of the new millennium, Sligar was also working with membrane proteins in his lab and growing frustrated. So his “wild idea” was to make a mixture of scaffold protein, lipid, and the membrane protein, and hope that in the process of self-assembly, the membrane protein would find itself a natural home inside the lipids. It worked! So Sligar then decided to hone the nanodisc assembly process, and provide the recipe for others wanting to study membrane proteins in their natural environment. In 2013, he offered an historical overview of the reactions to his initial nanodisc idea:
While the public response to this bio-assembly system was initially somewhat muted due to concerns that deeply embedded and totally integral membrane proteins would not readily be incorporated, subsequent efforts over the next decade (including that by many independent laboratories) demonstrated the broad applicability of nanodiscs for studying the breadth and complexity of membrane protein complexes.
While Sligar was finessing the recipe for making nanodiscs, around 2004, there was a graduate student rotating in Sligar's laboratory who told Sligar she was also interested in the computational side of biology; Sligar effectively sent her to work with Schulten for most of the rotation, and a collaboration between Schulten and Sligar's group was born. In the middle was graduate student, Amy Shih, who earned a BS in biochemisty and an MS in computer science before moving to Urbana. For her project on the nanodisc, she had to take a hybrid approach and balance a mixture of experimental, computational, and theoretical demands while she was puzzling out how to visualize these novel particles. “I've always enjoyed both computational work as well as experimental work,” says Shih. Fortunately for Sligar and Schulten, Shih had training in experimental biochemistry, and had done extensive computer coding as well, two skills demanded of her as she had to bridge two groups and two advisors. But after working a short time for her rotation with Schulten and Sligar, Shih fell in love with the nanodisc project assigned to her.
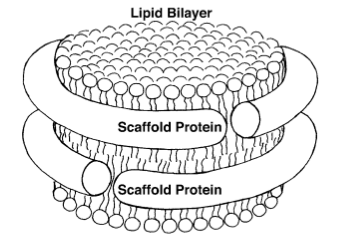
A schematic image of a nanodisc, a bioengineering device developed at the University of Illinois by Steve Sligar.
Sligar effectively needed Schulten's computational microscope to verify the shape of the nanodisc. While Sligar had done extensive experiments to determine a rough shape of the nanodiscs, he couldn't generate a high-resolution structure of his creation. He knew roughly that a nanodisc was about 10 nanometers wide, but this is much too small to see under a light microscope. And cryo-electron microscopy requires freezing. Schulten says Sligar used his atomic force microscope with the nanodisc on a Teflon substrate to get a rough image as a starting point. “It wasn't clear that under these conditions,” Schulten says of the atomic force microscopy, “that the disks are really still natural or organize themselves the way they do when you have them normally prepared.” It was really a perfect job for the computational microscope to step in.
One of the first things Sligar wanted Schulten's help with was to determine what were the bare essentials needed of the membrane scaffold protein so that nanodiscs still formed. It was noted in experiments that even as the scaffold proteins were systematically truncated, the result was always identical, 10 nanometer-wide particles, with 80 lipids on top and 80 lipids on the bottom, and two proteins encircling the assembly, as shown in the schematic drawing. “So they were particularly interested in optimizing how much of that protein they actually needed to make their base, 10 nanometer nanodisc,” Shih explains. And Schulten and Shih could actually see the reason with molecular dynamics simulations. The first eighteen or so residues of the scaffold proteins were not involved in binding the lipids.
It is not actually straightforward to run a molecular dynamics simulation if your particle has no accompanying high-resolution structure. And at the time Shih and Schulten wanted to do their simulations, around 2004, no high-resolution structure yet existed, so they fashioned one computationally. But Schulten had plenty of experience modeling the HDL particles of pioneer Ana Jonas in 1997, and understood all too well the nuances of building apolipoproteins around lipid bilayers. Shih says that the extensive characterization of nanodisc shape done formerly by Sligar's group was a great guide when building the structure. One of the things that Schulten knew for certain was that the membrane scaffold proteins formed long alpha-helices, and most likely formed two belts around the bundle of lipids, much as a belt holds up trousers. With years of acquired knowledge from Schulten and Sligar, the lack of a high-resolution structure was not a total impediment to running molecular dynamics simulations.
Perhaps the best test of molecular dynamics, and his computational microscope, was when Schulten suggested they try to simulate the actual self-assembly process of the nanodisc. In their original molecular dynamics simulations mentioned above, the maximum time they simulated was about 10 nanoseconds, because that is what the computers and software at the time could manage. But the actual self-assembly process in the laboratory of Sligar was more like microseconds. That requires about 10,000 nanoseconds of simulation time–a seemingly impossible feat. What was Schulten thinking? How could they get around this limitation?
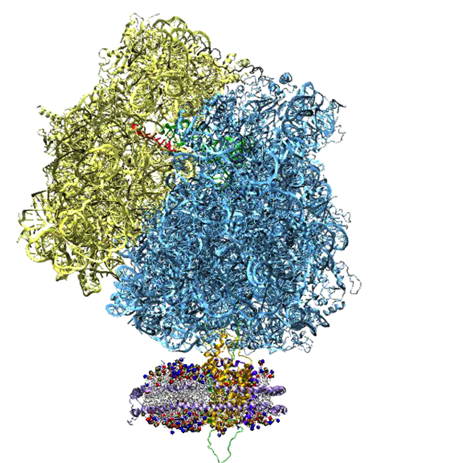
The ribosome on top, and the nanodisc on bottom, as Roland Beckmann imaged with cryo-electron microscopy.
The answer was: coarse-grained molecular dynamics. Instead of the software looking at every individual atom, groups of atoms were designated together as “beads,” cutting down the number of particles considered, thus reducing the fine details necessary in regular simulations, and effectively speeding up the process. Amy Shih says she actually worked for almost two years implementing coarse-grained molecular dynamics into NAMD. With such efforts, and only a small hint of payoff (another group had once shown lipid self assembly with coarse-grained molecular dynamics), Shih saw the lipids form into a disk, but the membrane scaffold proteins were just lying atop the structure. After two years of hard work, would the end result be non-ideal and disappointing? But Schulten had an idea: simulate for an even longer time. And that proved to be the solution. Shih says she was even surprised herself when it worked so well. The simulations, after 10 microseconds, showed a vivid portrait of the nanodisc forming and very much resembling, in its final form, the schematic image shown above. (See a movie of the nanodisc self-assembly process here.)
Not only did the scientists in Urbana see a vivid portrait of the nanodiscs forming, Shih drove up to Argonne and confirmed the coarse-grained molecular dynamics structure by doing small-angle X-ray scattering experiments. This was a wonderful triumph of the computational microscope, and really helped cement for interested researchers a correct image of the nanodiscs they were formulating in their laboratories to help elucidate membrane proteins.
But this story has a compelling postscript. Around 2009 Schulten was in Munich, visiting collaborator Roland Beckmann; the two were discussing their project on the ribosome, and Beckmann took Schulten out to look at a poster in the hallway. Schulten immediately saw in the poster an image of the nanodisc, and he was astounded! Roland Beckmann did not say a word, but Schulten immediately recognized the nanodisc he had already painstakingly visualized for the world to see some five years earlier. And there it was, in a cryo-electron microscopy image! “We had developed this as a tool, but we didn't think we would use it ourselves,” clarifies Schulten. “Many people did experiments, but this was the nicest experiment ever done with them. And Sligar and I didn't even know about the experiment.”
What Roland Beckmann had done was create a system of the ribosome plus the newly forming protein it was synthesizing, and this nascent protein was entering a protein-conducting channel living in a nanodisc. While Beckmann had taken an electron microscopy image of this four-part complex, he wanted Schulten's help in creating a high-resolution structure of it. Schulten and his team had just developed a method to fit high-resolution data (say from X-ray diffraction or NMR) into cryo-electron maps that are necessarily of lower resolution. Schulten's method is called Molecular Dynamics Flexible Fitting and a complete history of MDFF is found here, including a full explanation of Schulten's work with Beckmann on this novel undertaking. This system of Beckmann's was a great breakthrough for Sligar's method, and for Schulten it was a wonderful moment as well. Not only would he get to revisit the nanodisc in a new project that utilized the nanodisc in such an innovative fashion, but it was a complete verification of the image of the nanodisc structure that Schulten's computational microscope offered to the world.
Amorphous Silica–The Atomic Details
By the end of 2005 Schulten and Aksimentiev were fully immersed in studying biomolecules interacting with nanopores. They had studied nanopores in Greg Timp's silicon nitride membranes, and nanopores of alpha-hemolysin, which were naturally occurring in lipid membranes. Schulten and Aksimentiev realized a promising material for fabricating nanopores to use in translocation of DNA, and therefore ideal for sequencing, might be amorphous silica. But they wanted a top-notch description of this silica interacting with water and DNA, for that is critical to the success of molecular dynamics simulations. At this point, Aksimentiev accepted a job as a full-time faculty member in the physics department there in Urbana, so the job of describing silica fell to Schulten's graduate student Eduardo Cruz-Chú. Luckily he was more than capable, because he didn't realize what he was getting into.
Cruz-Chú had always loved chemistry but had a fascination with mathematics. He actually did experimental biophysics at the City College of New York before arriving in Urbana for graduate school. Once there, he said working for Schulten was a natural step as Schulten was very mathematically and computationally oriented. Cruz-Chú was a second or third year graduate when he started his project on silica in 2005. At this point in time, the field of bionanotechnology was really starting to gain a foothold in the world. It looked like combining biological macromolecules with silica would have several applications, such as nanoelectronics, self-assembly of nanostructures, microfluidics, DNA microarray technology, and nanopore sensors. What Schulten and Aksimentiev suggested was that Cruz-Chú develop an accurate description of how water and silica interact, namely, create from scratch a silica force field. At that point, silica, an inorganic material, had been commonly used in microelectronics as well as in semiconductor circuits to isolate different conducting regions. So if water got on these chips, manufacturers would have to throw them away! Therefore, while silica was well characterized, silica and water described together was a bit uncommon. It was a story waiting to be told.
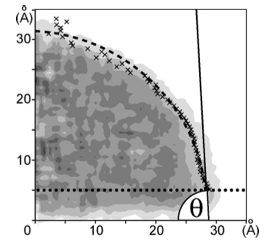
The water contact angle.
A force field describes the interactions between bonded and non-bonded atoms in a system. In essence it is a mathematical description for the potential energy of a system (in this case, silica plus water plus biomolecule). Most who work in this field, determining what are called force field parameters, will tell you that parameterization is a tedious task requiring multiple inputs, such as geometry optimizations, harmonic frequency calculations, and electrostatic potential fitting. But the quality of a molecular dynamics simulation often depends on the quality of its underlying force field, and Cruz-Chú was given the task of making sure the interaction between silica and water was properly parameterized.
Indeed, Cruz-Chú did not realize that his force field parameterization would present challenges even above and beyond the usual ones found in force field generation. “In modeling,” Cruz-Chú says of his force field work, “we develop models. And then these models aim to represent reality. But these models are in the end just models–they are representing one narrow part of reality. So this is the biggest problem when you parameterize, not only for me, but for anyone.” Cruz-Chú says the key to this problem of model building is validation. In this case he needed to find an experimental observable for his model to reproduce. And the answer came from Aksimentiev, who suggested that Cruz-Chú use the water contact angle.
The water contact angle is the angle between the tangent of the water droplet and the silica surface, and is shown in the image with an example. While it was a great idea to chose the water contact angle for validation of the silica force field, and would eventually make the publication very highly cited, Cruz-Chú said it was extremely difficult to implement. But he had a bit of luck that allowed him to succeed. Two publications came out at the same time, and truly propelled his project forward, according to Cruz-Chú. One article was about the parameterization of the water contact angle for carbon nanotubes. And another was a silica force field paper by Alex MacKerell. With those papers now as guidance, Cruz-Chú was able to tune his force field to accurately represent water on silica surfaces. While this silica project was certainly challenging for Cruz-Chú on so many levels, it produced many rewards, and it turns out Schulten and Aksimentiev's suggestion of parameterizing silica plus water was a good one. “Indeed, I always receive requests from that paper,” says Cruz-Chú. “Somebody wants the structure, or somebody wants a parameter file.” Cruz-Chú says he is very pleased to have been a second or third year graduate student and produce such a highly cited paper. “It was really quite a remarkable piece of work that he achieved,” summarizes Schulten.
While many other people have wanted to use the silica force field generated in Schulten's group, Cruz-Chú also used it in a further project he did with Aksimentiev and Schulten. The phenomenon they elucidated was the perfect candidate for the computational microscope.
The surface of the silica pore matters greatly for ion flow. On the left the ion (in blue) is in a raw cut pore, versus an annealed pore on the right.
As mentioned previously, when studying DNA translocating through a nanopore, the salty water that is also in the system is an important component to consider, for the currents in the nanopore are crucial indicators of behavior. While the overall goal was to examine DNA translocating in silica nanopores, an intermediate step is to consider the flow of just the ions in the pore as well. So Cruz-Chú started to examine the ionic flows in his silica nanopore. “And then the simulation showed the rectification,” Cruz-Chú notes. “I would say, when you have a good model, things just show up.” What Cruz-Chú was seeing was that the flow of ions in one direction was not the same in the other direction when he switched the polarity of the voltage applied across the pore. In a wire in a house, the charge carriers are strictly electrons, so the current flow is the same in the wire if one switches voltage polarity. But the charge carriers in the nanopore are things like calcium ions and chloride ions, and hence not uniform. Cruz-Chú was seeing rectification, or, that current flow in one direction was easier than the other direction. “It's the perfect case for molecular dynamics,” says Cruz-Chú, “because the size is small. And the atomic details matter a lot. Indeed, nanopores, they are so small, that EVERYTHING has an effect, I mean every water molecule, every ion, every little defect at the surface has an effect.” And Cruz-Chú could pick it all up with the computational microscope
In the simulations Cruz-Chú could see that what was happening at the surface of the nanopore determined how the ions were going to travel; the atomic details at the surface made the ions go faster or slower. Furthermore, he saw that defects at the surface were capable of removing water surrounding the ions that were retained by the surface. “Removing the water from the ion was something important,” reiterates Cruz-Chú. “You need the atomic description of the water. So this you cannot see with continuum modeling; you need every single water molecule to see what is happening.” Not only did the simulations show the importance of the surface of the nanopore, but Cruz-Chú, Schulten, and Aksimentiev decided to purposefully introduce defects at the surface to characterize subsequent rectification. So in the course of trying to study DNA in silica nanopores, the phenomenon of rectification showed up and the computational microscope was on hand to explain this at the atomic level, and led to an eventual publication.
One Small Group of Atoms–One Big Effect
Little did Klaus Schulten realize, when Greg Timp asked him in 2008 to take up a new project on methylated DNA, that this would launch Schulten's entry into epigenetics–a field that would soon be tagged as hot–or that he would go on to explore it in such an unconventional manner. Many scientists now study epigenetics, but there is so much left to uncover and explain, and so many opportunities for groundbreaking discoveries as well as cures for cancer and other diseases.
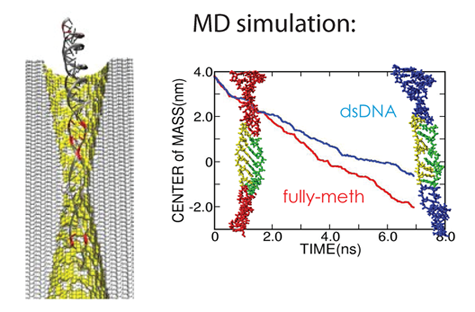
Simulations showed methylated DNA translocated nanopores more rapidly than its non-methylated cousin, corroborating experimental observations.
The crux of epigenetics is that there is a mechanism which alters an organism's DNA, but in such a way that the DNA sequence is left unchanged. Hence, the structure of the DNA is affected by this change, but the underlying sequences of G's, A's, T's, and C's are not. And this change is simply that a little group of atoms (a methyl group) is added to the cytosine (base C) in a DNA strand. It turns out that addition of this methyl group in strategic places can have huge consequences for humans, plants, and insects. And science is only beginning to figure it out.
One thing scientists are pretty certain about is that methylation of DNA is one way eukaryotes control gene expression, by turning off specific genes. But all the exact mechanisms are still being uncovered. Methylation patterns definitely are important for cellular processes. For example, methylation can actually silence tumor suppressor genes, and thus lead to certain cancers. Already there are drugs now that can target these methylation patterns and return the DNA to its non-methylated state and thus improve the lives of cancer patients.
Perhaps a fascinating illustration of the role of methylation comes from honeybee colonies. The three types of bees–workers, drones, and queens–all contain the exact same DNA, but their roles in the hive, and even their physical body shapes, are all different. Male drone bees are clones of the queen, but certain genes in these drones are turned off by methylation, and drones grow up to look and act differently from a queen bee. Scientists are pretty sure they understand the mechanism here. An egg destined to be a queen is fed an exclusive diet of royal jelly, and in this foodstuff there is a compound that inhibits the enzyme that methylates DNA. So queen bees have no methylated DNA, and hence are significantly morphologically different than workers or drones. This is a fascinating instance of epigenetics at work in the insect world.
In humans, scientists have found that methylation differs from human to human, for even identical twins don't have the same patterns of methylation. Diet, exercise, even environmental factors like war or psychological factors like fear, can change a human's level of methylation. The sequence of DNA in humans, for the most part, does not change during the course of a lifetime–the gene that codes for blue eyes won't suddenly change and make a person have hazel eyes; methylation patterns, however, change in the time between birth and death–they are different as a child, as a developing young adult, and then as an adult, and into old age. Recently scientists realized that they could use methylated DNA in healthy and cancerous tissues and cells to develop a sort of biological clock to chart the aging of different tissues in the human body. For example, breast tissue appears older than other tissues in a woman's body, and some cancerous tissues sped up the clock significantly. But as with many things in the field of epigenetics, what remains uncertain is if the epigenetic changes in DNA cause the ageing, or are caused by ageing.
The methods in the laboratory to detect the presence of methyl groups on DNA are time consuming, and often times require large sample sizes. A diagnostic tool that could detect methylated DNA cheaply and quickly would be of extreme benefit to the medical community. In 2008 Greg Timp had started a project using methylated DNA in his nanopores; his long-term goal was to provide a tool for simplified detection of methylation. He translocated both methylated DNA and non-methylated DNA through his silicon nitride nanopores and detected a difference. He wanted Schulten's help in explaining why he detected this difference–what was the underlying mechanism at work?
At this same time, Xueqing Zou, a physics graduate student at a university in Beijing, got a fellowship from the Chinese government to study abroad for a year, and so she chose to work with Schulten in Urbana. While she had a bachelor's degree in computer science, and so was a natural fit in Schulten's computationally-oriented group, she was new to the field of DNA methylation, the project Schulten assigned her. But she made quick progress and would soon become an expert in simulating methylated DNA for Schulten, for he hired her out of graduate school to then be his postdoc. Together, Schulten and Zou would grow more and more fascinated by epigenetics, and as the years progressed, through chance find themselves with projects where they could help explain the microscopic details to experimentalists designing novel ways to look at methylation.
The first experimental team Zou and Schulten worked with was Greg Timp's. He had found, surprisingly, that methylated DNA passed more easily through a nanopore than normal DNA. The simulations that followed showed that methylation changes DNA's ability to stretch itself through a nanopore. Methylated DNA is more compact, more ordered than its non-methylated cousin. “Non-methylated DNA is more flexible,” explains Zou. “It has more chance to interact with the wall of the nanopore, which will slow down the translocation velocity of non-methylated DNA.” Once again, as with the rectification work in silica nanopores, the molecules interacting with the surface of the nanopore played a big role in explaining the observed phenomena. These kinds of interactions, for example, of DNA atoms with surfaces, can only be illuminated with the computational microscope.
In 2012 an Urbana friend of Xueqing Zou, named Jiwook Shim, contacted her and said he and his advisor were doing experiments with methylated DNA in nanopores, and since he knew she had simulated these kinds of things previously, maybe she and Schulten could work with his group. The experimental group in question was run by Rashid Bashir, who headed the aforementioned Micro and Nanotechnology Laboratory in Urbana and is now head of UIUC's Bioengineering Department. Schulten was very eager to work with Bashir, and so a collaboration was born. While Zou and Schulten had already worked on methylated DNA in nanopores, Bashir's experiments were slightly different. His group compared non-methylated DNA with methylated DNA that was tagged with a binding protein, and they ran experiments with different pore sizes.
What the experiments determined, and what the simulations of Zou and Schulten confirmed, was that the pore size is key. As in the other nanopore experiments mentioned previously, the experimentalists looked at the blockade current to distinguish the objects undergoing translocation. When the pore diameter was too big, the current signals were covered by noise and it was impossible to distinguish the two types of DNA; but if the pore was too small, the DNA plus protein couldn't move through it at all. The computational microscope could show exactly what parts of the protein were interacting with the pore surface, and explain, for example, why the complex of methylated DNA plus protein got stuck in a small pore. Schulten and Zou's simulations also elaborated how binding sites between the protein and DNA could be ruptured depending on pore dynamics.
While Schulten and Zou worked with methylated DNA in nanopores for two different experimental groups on the Urbana campus, they would also embark on a completely different odyssey with an off-campus scientist to elucidate methylation, and meet with many roadblocks along the way.
The Mechanical Properties of Methylated DNA
As Klaus Schulten's collaborations with Urbana faculty have played a big role in this history, and his very locality on the campus offered him a plethora of opportunities to employ the computational microscope in biotechnology, another city is important in his career–Munich. And Munich presented him with the opportunity to extend his nascent work on epigenetics into a realm where literally no one else had traversed.
While Schulten left Munich in 1988 for the United States, he has maintained close ties with his former home, and spends a month or two every summer there, nurturing collaborations and keeping up with friends. He and his wife even manage to take advantage of the beautiful hiking afforded them in the Bavarian state. One of his friends there is Hermann Gaub, whom Schulten knew when Gaub was a doctoral student in the 1980s in physics at the Technical University of Munich, where Schulten was employed. Schulten stayed in contact with Gaub over the years, as Gaub was a professor first at the Technical University of Munich and then moved in 1995 to the Ludwig Maximillian University, also in Munich. Schulten followed Gaub's progress as Gaub became an expert in the field of mechanical forces in biology. But for twenty years the two scientists never worked on a project together. “You need to have a scientific reason to collaborate. So you must complement each other, and each party must need the other party,” says Schulten. “But the way that happens is actually often based on friendship, on knowing people for a long time, and trusting them, and so on.” When Gaub and Schulten talked about Gaub's novel measurements on methylated DNA in 2010, the duo realized that, after all this time as friends, they finally had a reason to collaborate.
The experiments Gaub was conducting on methylated DNA were based on a new technique he was developing to better study the binding forces in protein-DNA complexes. He calls this molecular force assay. The way it works is the following: Two strands of DNA are connected to two separate chips or surfaces. A target duplex is attached to the bottom chip, and a reference duplex is attached to the top chip. Then these two strands, target and reference, are coupled together in series. The chips each contain about 10,000 strands of target + reference DNA. The chips are then separated, and as the DNA strands rupture, a fluorescence marker indicates which broke, the target or the reference strand.
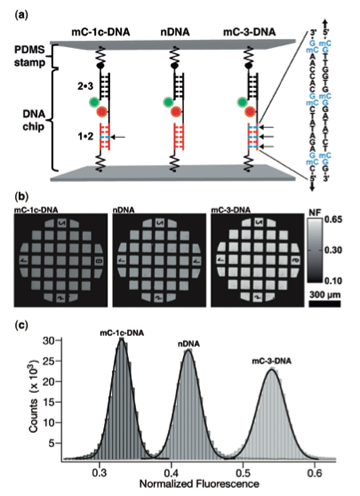
Studying methylated DNA with Molecular Force Assay, the brainchild of Hermann Gaub. The reference strand is in black while the target strand is in red.
With the fluorescent read-out, it is possible to get a distribution of the results of the strands rupturing. If the target DNA and reference DNA are exactly the same, one would expect that the top strand ruptures about 50% of the time, and the bottom strand ruptures about 50% of the time. But the genius of this method is it makes around 10,000 measurements simultaneously, ALL UNDER THE SAME CONDITIONS. This is critically essential when one is looking at such a small system like a piece of DNA, for the molecule is highly subject to thermal fluctuations. So this experimental apparatus provides extreme measurement sensitivity.
Another key aspect to the molecular force assay is that it is a comparison measurement. Schulten likes to point out that one of the most important things he learned in his first physics class as a young boy was that measuring means comparing. To weigh out a pound of sugar on a balance scale, a one-pound weight is put on one pan, and then sugar is slowly added to the other pan until the two pans are equally balanced–this is a classic example of comparison measurement. Gaub, in molecular force assay, can make up to 10,000 comparison measurements at once.
To study epigenetics, Gaub, with graduate student Philip Severin, looked at molecular force assays with the lower duplex (in red) having either zero, one, or three methyl groups added to cytosines in the DNA strand. The top strand, or reference duplex, as expected, had no methyl groups added at all. When they did the experiments, they observed the distribution curve shown. Enter Schulten and the computational microscope, to help explain the underlying physical mechanism of the assay results. Xueqing Zou worked on the simulations of the methylated DNA for this project as well. “In our simulations,” says Zou, “we observed that DNA methylation patterns can influence the mechanics of DNA.” What Zou and Schulten noted with molecular dynamics was that when DNA gets stretched, bubbles develop in the DNA, and these bubbles mean the base pairing in DNA is broken. As DNA is stretched, bubbles develop, and the more bubbles that develop, the less stable the DNA is. What the team in Urbana saw was that methylated DNA develops fewer bubbles than non-methylated DNA, hence methylated DNA is more mechanically stable.
Despite this novel experiment on methylated DNA, plus the underlying physical mechanism explained with molecular dynamics, the Munich-Urbana team had extreme difficulties publishing. The first two journals they submitted their manuscript to rejected them outright. While other groups had looked at the thermal properties of methylated versus non-methylated DNA, and had detected very little difference, and another group had studied the flexibility of methylated DNA, no on had ever looked at the mechanical property of methylated DNA in quite this way. Schulten believes the referees thought this mechanical property had nothing to do with epigenetics, and so rejected the manuscript because of that. But then a finding came out, which Schulten believes helped their case and helped them eventually publish in Nucleic Acids Research.
In the sum total of human DNA, it is predicted that only about 2% codes for proteins. The rest is labeled as non-coding DNA, which was once referred to as “junk DNA.” In 2012 a team of scientists sequenced all the non-coding DNA in the human genome, and found that it is highly methylated. As mentioned above, methylation is one form of controlling gene expression. But why would non-coding regions be methylated? “If epigenetics methylates something, that's a costly process, they are not doing that just for the fun of it,” reiterates Schulten. “But if it's not the reason of controlling directly the DNA, there must be some other reason.” Schulten postulates that because DNA is on the chromosome, and the chromosome is highly wound and needs to be unraveled to get at the DNA, that non-coding DNA likely has an attribute related to its mechanical properties. “And then suddenly what we found was very interesting,” says Schulten of the mechanical properties of methylated DNA elucidated by Gaub's experiments and Schulten's simulations. “This was one of the first very concrete indications that methylation actually affects the mechanical properties of DNA,” notes Schulten. “And that makes a lot of sense in the context of non-coding DNA being very often epigenetically modified.”
Graphene and DNA Translocation
While many scientists are working furiously to find a cheap and swift way to sequence DNA through nanopores, the ultimate answer might come from a novel material called graphene. Schulten has teamed up with an electrical engineer also at the Beckman Institute to illuminate graphene nanopores.
Graphene was discovered in 2004, and the scientists who found it won the Nobel Prize in 2010. This material is basically a thin sheet of carbon atoms living in a honeycomb lattice. It is stronger than steel, and conducts electricity better than copper. The reason graphene shows such promise in the field of nanopore sequencing is that it is basically a monolayer thick. The distance between subsequent base pairs of DNA is about 0.35 nanometers. A typical solid-state nanopore is about 10-20 nanometers thick–which means that many bases of DNA remain in the nanopore and the current blockade signature reflects the profile of many bases at once. But graphene's thickness is comparable to the distance between base pairs, meaning roughly one base translocates through a graphene nanopore at a time. Therefore, with a nanopore whose membrane is about as thick as the base-to-base stacking distance of DNA, the resolution of the DNA bases should be subsequently better. Unlike its semiconductor counterparts, the sheet of monolayer-thick carbon also can conduct electricity tranversely, which might prove useful in nanopore sequencing as well. Thus, with all its proposed uses, graphene as DNA sequencer holds promise.
Electrical engineer Chaitanya Sathe joined Klaus Schulten's group as a graduate student in 2009 and was looking for a promising project in nano-engineering. One day he was talking with Xueqing Zou, who, as we met earlier, had done many projects in methylated DNA translocating in solid state nanopores, and they started talking about graphene and the new experimental papers coming out demonstrating double stranded DNA translocation in graphene nanopores. Sathe and Zou decided this would warrant theoretical descriptions, and Sathe had his promising project. Schulten and Sathe then teamed up with fellow Beckman scientist Jean-Pierre LeBurton to look at graphene nanopores under the computational microscope.
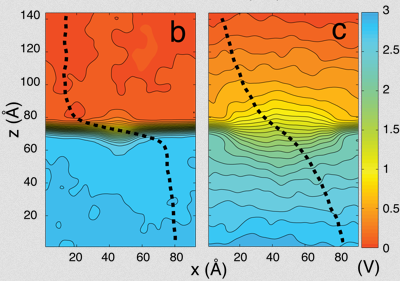
The electric field inside the graphene nanopore depends on pore diameter. On the left is a 2 nm pore, and on the right is a 7nm pore.
One of the first things that unexpectedly showed up in the simulation of nanopore, water, and ions was what the Beckman scientists called the breathing of the membrane. The membrane actually jumps wildly up and down, exhibiting thermal fluctuations easily picked up by molecular dynamics. Additionally, Sathe used many of the protocols outlined by Aleksei Aksimentiev and Schulten in their “Imaging alpha-Hemolysin” publication, and came up with his own “images” of graphene nanopores. “Perhaps what was really interesting,” explains Sathe, “that we observed from our simulations, is that because this graphene layer is so thin, we could tailor the electric field in the pore with very great accuracy.” Depending on the pore diameter, the electric field behaves differently inside the pore, as can be seen in the potential map of a 2 nanometer versus a 7 nanometer pore.
The ultimate goal of DNA sequencing is to detect with high resolution whether an A, T, C, or G is going through the pore. But an intermediate step in honing the graphene nanopore sensor is to consider long double strands of just As paired with their counterpart Ts, and, then, alternately, strands of just Gs paired with Cs. The scientists at Beckman did extensive studies of A-T versus G-C strands to demonstrate how a nanopore might differentiate between the two different types of strands.
The applied voltage across the pore translates, as we've seen before, to DNA in the pore getting stretched by the resultant forces from the electric field in the pore. “When the force is too large,” explains Sathe, “actually both A-T and G-C stretch to the same degree. And you see that there's no difference in the ionic current.” However, there is a window where one strand stretches more than the other, as A-T strands stretch more readily because they have one less intermolecular hydrogen bond than G-C. And so it would be possible to use this window of operation of the bias voltage to actually detect A-T versus G-C. Sathe says this is one of the most exciting results of the graphene nanopore work, which was published in ACS Nano in 2011. It is a first step in demonstrating the feasibility of graphene to detect at the sensitivity level of a single base pair. For Klaus Schulten and Aleksei Aksimentiev, who in 2011 rejoined the Theoretical and Computational Biophysics group as affiliate faculty after a seven-year hiatus, it is a testament to the protocols the two scientists pioneered in their initial studies of alpha-hemolysin and solid state nanopores in 2005. These protocols are now being employed by both the Schulten and Aksimentiev groups regularly, and the rest of the world is starting to take notice.
While the predictive power of the computational microscope was harnessed to suggest the feasibility of the graphene nanopores for DNA sequencing, Schulten's years of work in bionanoengineering would culminate in a project that not only used the computational microscope to explain the underlying physical mechanism, but also helped guide a bioengineer to totally rethink his nanosensor design.
A New Way of Conducting Biology Research
Professor Logan Liu, who works at the Micro and Nanotechnology Laboratory in Urbana, was at a standstill with a new biosensor he was developing to detect kinase activity. So he decided to approach Klaus Schulten to see if Schulten could help illuminate what might be going on with a simulation. Little did Liu realize that he would change the way he conducted his bio-detection work by consulting with Schulten who worked, conveniently enough, just down the street.
Logan Liu's kinase sensor. When a positive voltage is applied to the gold nanoparticles (GNPs), the peptides move closer to the surface of the gold.
A living cell is continually carrying out numerous functions just to survive. Some common examples of cellular processes are transporting molecules, making proteins, converting nutrients into usable energy, and reproduction. Because a cell is a complex machine, it needs to coordinate processes–initiate them or stop them–and there are so-called pathways where proteins of the cell send signals to one another to coordinate the necessary processes. Each signaling pathway can consist of a few to a multitude of proteins that are somehow linked to each other, with the express purpose of completing a task within a cell. A rough picture of this kind of signaling is to think of a domino effect, where one protein signals the next then the next, and so on, and at the end the result might be that a gene gets expressed. These signaling pathways are often complex and a scientist may spend years just figuring out one complete signaling pathway.
But how does one protein send a signal to another protein? Enter the kinases. They are a type of protein that is involved in almost all cellular processes. And the kinases generate a very important signal, namely, they phosphorylate another protein (they add a phosphor group to a specific amino acid on the receptor protein). Basically kinases are turning switches on and off by adding or subtracting phosphates to the next protein in the pathway. Their importance is highlighted by the fact that there are well over 500 protein kinases in the human body. It has also been recognized that kinases are involved in cancer and disease, where a kinase may be too active, or not active enough. For example, several mutations in the kinases have been implicated in non-small-cell lung cancer, and advances have been made with targeted therapies that specifically inhibit the kinase activity, and lead to prolonged life spans for patients. Suffice it to say, kinase activity in humans is critical from so many angles.
Logan Liu, who got his PhD in bioengineering in 2006, has been thinking about kinase activity since his graduate school days at University of California, Berkeley and University of California, San Francisco, and then again in his postdoc at the Helen Diller Family Comprehensive Cancer Center in San Francisco. “In my research, we're interested in studying this from a systematic level,” Liu says about the kinases, “which means from the family-of-proteins level. We want to study the relationship between not only one but multiple kinase enzymes, and their responsivity, as well as activities in different kinds of cell lines, and the same cell line, in different stages of cancer development.”
By 2010, now at the University of Illinois, Liu had developed a preliminary nanosensor to detect kinase activity, but he wasn't quite getting the results he expected. An expert in bionanotechnology, Liu had created his device using gold nanospheres, working on the premise that biomolecules like proteins or DNA behave in certain ways when placed on surfaces or substrates (compared to how they behave when floating in an aqueous environment, as they are in a cell).
His original idea of a kinase sensor was the following: Onto a gold nanosphere, place a peptide segment and in the middle of this segment have a tyrosine, because this is often the target amino acid that gets phosphorylated by the kinases. Because a phosphate group is negatively charged, when a phosphor attaches to the tyrosine, and a positive voltage is put on the gold, the peptide segment should move down toward the surface. To detect this movement to the surface, he placed atop the peptide probe a molecule that can fluoresce, a laser dye called rhodamine. Thus the complex, of protein segment plus phosphate plus rhodamine, when it moves to the surface of the gold, should show surface fluorescence enhancement. All this is based on the fact that the peptide probe will be at different distances from the gold surface depending on whether or not it has an attached phosphor, and the signal from the dye molecule should reflect this.

The computational microscope, a tool honed by Klaus Schulten's group at the Beckman Institute. This tool is often the only way to see the atomic details of bionano-devices in action.
But Liu wasn't getting the results he expected and so asked Schulten if Schulten's group could simulate his experimental setup, to help him explain what might be going awry. At this point he saw the role of the computational biologist as helping to explain and verify experimental results. It was the summer of 2010 and Schulten had just hired an undergraduate from Oberlin, Jaie Woodard, to work at Beckman. He paired her up with senior graduate student Eduardo Cruz-Chú, although Woodard already had experience using NAMD for molecular dynamics simulations she had run in her time at Oberlin. The pair set to work on the kinase system.
In the initial experiment, the apparatus containing the gold nanospheres looked like a Christmas tree. That is, a small conical surface held dozens of gold spheres, so that they resembled ornaments. And the gold held a whole lawn or protein segments. “When we simulated the system, we saw that the rhodamines from different peptides would often aggregate or stick together,” recalls Woodard. The result was that the rhodamines, very flat molecules, stuck together like glue and did not really move much when a positive voltage was applied. To make an effective sensor, the rhodamines had to come significantly closer to the surface than what the simulation was showing. They had solved Liu's mystery. Not only were the rhodamines aggregating, which made the complex unable to move much when a positive voltage was applied, but it looked in the simulations like the rhodamine clump might also keep the kinase from accessing the tyrosine residue to phosphorylate it.
Liu now needed another signaling element, as the laser dye was no longer an option. After much debate and discussion, Liu decided to try another spectroscopy: surface-enhanced Raman Spectroscopy. When a molecule absorbs and then re-emits light in Raman scattering, if the molecule moves toward the surface, a signal enhancement can be noted. Thus a phosphorylated peptide near the surface should show a different signal than a peptide without a phosphate, which is necessarily farther from the gold surface. To see if eradicating the rhodamine would have the desired effect, the theorists put their computational microscope on the proposed system. In the new simulations, they saw the peptide bend toward the gold surface with an applied voltage. “In particular, we saw the negatively charged phosphate group approach the gold,” Woodard said. “And it was the change in the conformation of the peptide when we applied an electric field in the simulations that made us think this new approach would work.” Indeed, in the laboratory, the key was to remove the rhodamine and use the alternative spectroscopy. When Liu's group looked for a signal enhancement with an applied positive voltage, the experimentalists saw one.
But the power of the computational microscope did not stop there. It held amazing predictive value, according to Logan Liu. Schulten and his crew worked on optimizing the sequence of the peptide probe, tweaking it to get the strongest adhesion to the surface. For example, the simulations showed that the lysine groups near the gold surface were sticking to both the gold and to the other residues within the peptide. Viewing this kind of atomic detail to help craft a better sensor was only possible in simulation, and was not visible in the laboratory experiments. “We thought that by changing this lysine residue to a different amino acid we could decrease the amount of interaction with the gold and the other peptide residues, and we could possibly increase the flexibility of the peptide under an applied field,” summarizes Woodard. So they replaced the lysine with alanine. And the simulations showed the alanine did not adhere to the gold or other residues in the same way as the lysine, as they had predicted.
For Logan Liu, he really appreciated the help of the computational microscope, and grew to see its power as a predictive tool that could aid in experiment design, which he said for him is based on one part intuition, and another part exhaustive literature searches. “And what we also learned,” says Liu about working with Schulten, “is that we can actually do a simulation first and then use the simulation as a very efficient way and, I would say, an intelligent design way–rather than a trial-and-error method to guide our experiments.”
Liu likes the fact that with molecular dynamics, nothing is in blind sight, and he has some expectation of what is going to happen before he runs the experiment; he also likes that he can choose the optimum experimental conditions, such as the best peptide sequence or the ideal substrate surface. He says it totally changes the way he is doing some bio-detection experiments, and he hopes to foster a continuing collaboration with Schulten. “I saw a gap,” continues Liu, “between experimental biologists or bioengineers and the computational biologist. But here we are trying to mend the gap. We're trying to build a bridge between those two fields through our collaboration.” He hopes more experimentalists will recognize the value of the computational microscope in helping them design better experiments. As Liu says, “that may be a new way of doing biology research in future.”
Conclusion
In retrospect, since 2002 Schulten has had many successes in computational bionanotechnology. But it has not always been easy. Schulten entered this field in his mid-fifties, and had to contend with all the ramifications of being a novice in the nanotechnology discipline. “When you are working in a new area and you go to a conference, you feel really bad, because you are nobody,” he explains. “And when you are getting older and you are nobody, people think you are an idiot.” But Schulten recognizes this as natural, for it is part of growing in science–slowly making progress and slowly getting to know the people in the field.
While he was one of the first people to work on simulating biological molecules combined with nanodevices, his entry into this field at age fifty-five was just right–Schulten says he won't enter a new field unless he can be sure he has ten years to devote to it; he believes it really takes at least a decade to make an impact in science. So with retirement approaching, this was probably one of the last few endeavors he could responsibly tackle.
Part of the reason Schulten has made such headway in this field is because he is not shy about studying technology–he believes that technology encompasses about ten to twenty percent of his research schedule. And he feels comfortable taking on projects about biotechnology because he works at the Beckman Institute. There, science and technology go hand in hand; he does not have to listen to cries of his imminent intellectual downfall if he teams up with engineers. It is actually encouraged at Beckman to cross disciplines in this way.
Finally, while Schulten took a risk in using molecular dynamics to simulate systems of biomolecules plus nanodevices, it paid off in part because he had spent all of his professional life honing the computer to be an effective tool in biophysics. And when opportunities to use the computational microscope in bionanotechnology applications started presenting themselves, Schulten realized the computer could really contribute to guiding the engineer. The timing was perfect, in fact. Schulten had the right tools for the job, and was not yet near retirement. Schulten also realized that it was a unique moment in the history of science–technology was offering him novel opportunities for research. And while the computational microscope undergirds the science in this article, for it is just the right imaging technique for bionano-applications, Schulten himself recognized how valuable this computer technology itself could be in studying novel technological applications fabricated by his innovative collaborators. As he is fond of pointing out, technology also drives science.
Other Related Articles
If you enjoyed this article, you might be interested in these other histories:History of Magnetoreception
History of Polyenes
History of Membrane Proteins
History of Photosynthesis
History of NAMD
History of VMD
History of MDFF (Molecular Dynamics Flexible Fitting)
Comments?
For comments or inquiries, Lisa Pollack can be reached at the following: lpollack AT ks.uiuc.eduCitation
To cite this article, please use the following:Pollack, Lisa. "Bionanotechnology and the Computational Microscope." 18 Dec 2013, from http://www.ks.uiuc.edu/History/BioNano/
References
Theodore L. Brown. Bridging Divides: The Origins of the Beckman Institute at Illinois. Urbana and Chicago, University of Illinois Press, 2009.
J. Andrew McCammon, Bruce R. Gelin, and Martin Karplus. Dynamics of Folded Proteins, Nature, 267:585-590, 1977.
Eric H. Lee, Jen Hsin, Marcos Sotomayor, Gemma Comellas, and Klaus Schulten. Discovery through the computational microscope. Structure, 17:1295-1306, 2009.
Marcos Sotomayor, David P. Corey, and Klaus Schulten. In search of the hair-cell gating spring: Elastic properties of ankyrin and cadherin repeats. Structure, 13:669-682, 2005.
Gwangrog Lee, Khadar Abdi, Yong Jiang, Peter Michaely,Vann Bennett, and Piotr E. Marszalek. Nanospring behaviour of ankyrin repeats. Nature, 440:246-249, 2006.
Rosemary Braun, Mehmet Sarikaya, and Klaus Schulten. Genetically engineered gold-binding polypeptides: Structure prediction and molecular dynamics. Journal of Biomaterials Science, 13:747-758, 2002.
J. B Heng, A. Aksimentiev, C. Ho, P. Marks, Y. V. Grinkova, S. Sligar, K. Schulten, and G. Timp. Stretching DNA using an electric field in a synthetic nanopore. Nano Letters, 5:1883-1888, 2005.
John J. Kasianowicz, Eric Brandin, Daniel Branton, and David W. Deamer. Characterization of individual polynucleotide molecules using a membrane channel. Proceedings of the National Academy of Sciences, USA, 93:13770-13773, 1996.
Aleksij Aksimentiev and Klaus Schulten. Imaging alpha-hemolysin with molecular dynamics: Ionic conductance, osmotic permeability and the electrostatic potential map. Biophysical Journal, 88:3745-3761, 2005.
Jéröme Mathé, Aleksei Aksimentiev, David R. Nelson, Klaus Schulten, and Amit Meller. Orientation discrimination of single stranded DNA inside the α-hemolysin membrane channel. Proceedings of the National Academy of Sciences, USA, 102:12377-12382, 2005.
Deyu Lu, Yan Li, Umberto Ravaioli, and Klaus Schulten. Ion-nanotube terahertz oscillator. Physical Review Letters, 95:246801, 2005.
Mary A. Schuler, Ilia G. Denisov, and Stephen G. Sligar. Nanodiscs as a New Tool to Examine Lipid-Protein Interactions. In Lipid-Protein Interactions: Methods and Protocols, Methods in Molecular Biology, pp. 415-433, 2013.
Amy Y. Shih, Ilia G. Denisov, James C. Phillips, Stephen G. Sligar, and Klaus Schulten. Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophysical Journal, 88:548-556, 2005.
Amy Y. Shih, Anton Arkhipov, Peter L. Freddolino, and Klaus Schulten. Coarse grained protein-lipid model with application to lipoprotein particles. Journal of Physical Chemistry B, 110:3674-3684, 2006.
Eduardo R. Cruz-Chú, Aleksei Aksimentiev, and Klaus Schulten. Water-silica force field for simulating nanodevices. Journal of Physical Chemistry B, 110:21497-21508, 2006.
Eduardo R. Cruz-Chú, Aleksei Aksimentiev, and Klaus Schulten. Ionic current rectification through silica nanopores. Journal of Physical Chemistry C, 113:1850-1862, 2009.
Steve Horvath. DNA methylation age of human tissues and cell types. Genome Biology, 14:R115, 2013.
Utkur M. Mirsaidov, Winston Timp, Xueqing Zou, Valentin Dimitrov, Klaus Schulten, Andrew P. Feinberg, and Greg Timp. Nanoelectromechanics of methylated DNA in a synthetic nanopore. Biophysical Journal, 96:L32-L34, 2009.
Jiwook Shim, Gwendolyn I. Humphreys, Bala Murali Venkatesan, Jan Marie Munz, Xueqing Zou, Chaitanya Sathe, Klaus Schulten, Farhad Kosari, Ann M. Nardulli, George Vasmatzis, and Rashid Bashir. Detection and quantification of methylation in DNA using solid-state nanopores. Scientific Reports, 3:1389, 2013.
Philip M.D. Severin, Xueqing Zou, Hermann E. Gaub, and Klaus Schulten. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Research, 39:8740-8751, 2011.
Chaitanya Sathe, Xueqing Zou, Jean-Pierre Leburton, and Klaus Schulten. Computational investigation of DNA detection using graphene nanopores. ACS Nano, 5:8842-8851, 2011.
Yi Chen, Eduardo R. Cruz-Chú, Jaie Woodard, Manas Ranjan Gartia, Klaus Schulten, and Logan Liu. Electrically induced conformational change of peptides on metallic nano-surfaces. ACS Nano, 6:8847-8856, 2012.



