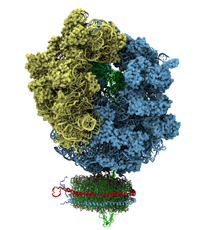
The activation of coagulation factor X (FX) is arguably the most important step in the formation of blood clots. When tissue is damaged by external injury, FX activation is usually initiated by the formation of a complex between FVIIa and tissue factor, which binds and activates FX after anchoring into cellular membranes rich in anionic lipids and calcium. This key step involves the formation of a tripartite complex on the membrane, which was earlier modeled by Resource researchers and published in Blood Advances. In collaboration with the Morrissey lab at the University of Michigan, the previous model is now confirmed by a new, and first, cryo-EM structure of the tripartite complex on the membrane, a study reported as a cover story in Blood.
The Future of Biomolecular Modeling
The Annals of MDFF
Announcements
Seminars
Remembering Klaus Schulten
Recent Publications All Publications
- LetA defines a structurally distinct transporter family. Nature, 2026.
- Simulating Gas Permeation Through the Central Pore of AQP5. Adv Exp Med Biol., 1498:105–113. 2026.
- Elevator mechanism dynamics in a sodium-coupled dicarboxylate transporter. Proc. Natl. Acad. Sci. U.S.A., 123(2):e2500723123. 2026.
- Impact of Lipid Composition on Membrane Partitioning and Permeability of Gas Molecules. Membranes, 16(1):33. 2026.
- Probing the role of membrane in neutralizing activity of antibodies against influenza virus. Structure, 34(2):254–263. 2026.
- Impact of anionic lipids on the energy landscape of conformational transition in anion exchanger 1 (AE1). Nat. Commun., 16:11664. 2025.
- Drug-bound outward-facing conformation of a heterodimeric ABC exporter suggests a putative mechanism of drug translocation. Nat. Commun., 16:10403. 2025.
Highly Cited
Click here for other highly cited papers