VMD-L Mailing List
From: Dr. Seth Olsen (seth.olsen_at_gmail.com)
Date: Thu Sep 21 2006 - 21:25:09 CDT
- Next message: Dr. Seth Olsen: "Re: What unit system does VMD use?"
- Previous message: Dr. Seth Olsen: "How to hide graphics molecules in a python script?"
- In reply to: Axel Kohlmeyer: "Re: What unit system does VMD use?"
- Next in thread: Dr. Seth Olsen: "Re: What unit system does VMD use?"
- Reply: Dr. Seth Olsen: "Re: What unit system does VMD use?"
- Reply: Axel Kohlmeyer: "Re: What unit system does VMD use?"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]
Hi Axel,
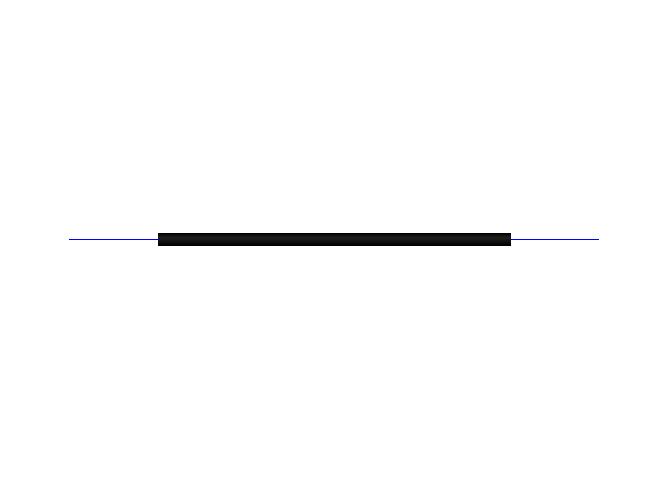
If this is the case, then something is definitely wrong. Attached please
find a python script named 'VMDAngstrom.py', a pdb file named '
AngstromScale.pdb', and a JPG file named 'Angstrom.jpg'. The pdb contains 2
nitrogen atoms placed at (-0.5,0.0,0.0) and (0.5,0.0,0.0). The script loads
this pdb (lines drawstyle), then creates a graphics molecule and draws a
thick black cylinder cylinder with endpoints at (-0.5,0.0,0.0) and (0.5,0.0,
0.0). The JPG shows just what I got when I ran the script.
I am running a precompiled linux binary of VMD 1.8.5. I have pointed
PYTHONHOME to a precompiled python binary (compiled for 1.8.4, but there is
no current distribution as far as I can tell). My OS is FC5 running on a
P4.
What's even more frustrating is that it seems that different results are
obtained if you create the graphics molecule, then load the pdb.
Regardless, I don't think this is the way VMD was intended to work.
Cheers,
Seth
On 9/22/06, Axel Kohlmeyer <akohlmey_at_cmm.chem.upenn.edu> wrote:
>
> On 9/21/06, Dr. Seth Olsen <seth.olsen_at_gmail.com> wrote:
> >
> > Hi VMDers,
> >
> > I've been writing python scripts to visualize 3N dimensional vectors (
> i.e.
> > molecular modes) as arrows attached to atoms. I've been having a lot of
> > problems with length distortions and after checking my code over several
> > times I think that the problem might be that I have assumed that VMD
> uses an
> > internal length scale that I am not. What are the units that VMD uses?
>
> all graphics objects are drawn on an angstrom scale.
>
> axel.
> >
> > Cheers,
> >
> > Seth
> >
> > --
> > ccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccms
> >
> > Dr Seth Olsen, PhD
> > Postdoctoral Fellow, Biomolecular Modeling Group
> > Centre for Computational Molecular Science
> > Australian Institute for Bioengineering and Nanotechnology (Bldg. 75)
> > The University of Queensland
> > Qld 4072, Brisbane, Australia
> >
> > tel (617) 3346 3976
> > fax (617) 33654623
> > email: s.olsen1_at_uq.edu.au
> > Web: www.ccms.uq.edu.au
> >
> > ccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccms
> > The opinions expressed here are my own and do not reflect the positions
> of
> > the University of Queensland.
>
>
> --
> =======================================================================
> Axel Kohlmeyer akohlmey_at_cmm.chem.upenn.edu http://www.cmm.upenn.edu
> Center for Molecular Modeling -- University of Pennsylvania
> Department of Chemistry, 231 S.34th Street, Philadelphia, PA 19104-6323
> tel: 1-215-898-1582, fax: 1-215-573-6233, office-tel: 1-215-898-5425
> =======================================================================
> If you make something idiot-proof, the universe creates a better idiot.
>
-- ccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccms Dr Seth Olsen, PhD Postdoctoral Fellow, Biomolecular Modeling Group Centre for Computational Molecular Science Australian Institute for Bioengineering and Nanotechnology (Bldg. 75) The University of Queensland Qld 4072, Brisbane, Australia tel (617) 3346 3976 fax (617) 33654623 email: s.olsen1_at_uq.edu.au Web: www.ccms.uq.edu.au ccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccmsccms The opinions expressed here are my own and do not reflect the positions of the University of Queensland.
- text/x-python attachment: VMDAngstrom.py
- chemical/x-pdb attachment: AngstromScale.pdb
- Next message: Dr. Seth Olsen: "Re: What unit system does VMD use?"
- Previous message: Dr. Seth Olsen: "How to hide graphics molecules in a python script?"
- In reply to: Axel Kohlmeyer: "Re: What unit system does VMD use?"
- Next in thread: Dr. Seth Olsen: "Re: What unit system does VMD use?"
- Reply: Dr. Seth Olsen: "Re: What unit system does VMD use?"
- Reply: Axel Kohlmeyer: "Re: What unit system does VMD use?"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]



