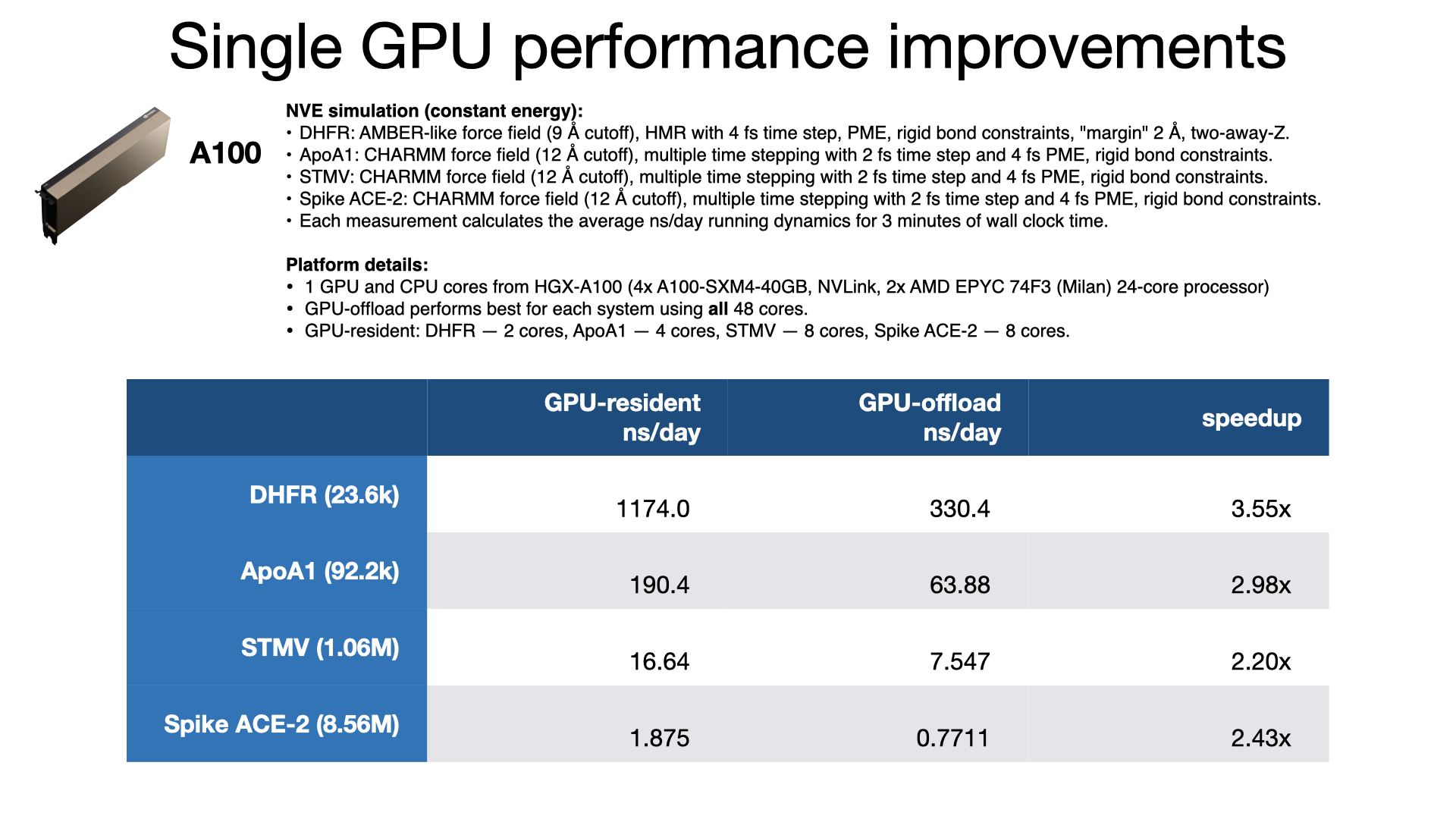
NAMD Performance
NAMD 3.0 beta 6 GPU-resident benchmarking results
NAMD 3.0 has a new GPU-resident simulation mode available, for multicore and netlrts builds, that maximizes performance of small- to medium-sized molecular dynamics simulations, enabling more than 2x performance gain over the earlier GPU-offload mode. In addition to performing force calculations on GPU, GPU-resident mode performs the numerical integration and rigid bond constraint calculations on GPU. The simulation data is maintained on the device between time steps, eliminating per-step host-device communication latencies. With very little work remaining on the CPU cores, the new GPU-resident mode no longer needs a large number of CPU cores to "keep up" with each GPU, which is reflected in the much greater aggregate throughput performance now possible on GPU-dense architectures.
The GPU-resident simulation mode is also capable of scaling a simulation across multiple GPUs in a single-process, shared-memory build of NAMD. Multi-GPU GPU-resident simulation requires a peer-to-peer connection between all pairs of GPU devices. Although this is possible to do over PCIe, good scaling performance requires a higher bandwidth, lower latency interconnect, like NVIDIA NVLink. Supported hardware platforms, like NVIDIA DGX, show GPU-resident achieving 7x performance gains over GPU-offload, due to the very poor single-node multi-GPU scaling of GPU-offload.
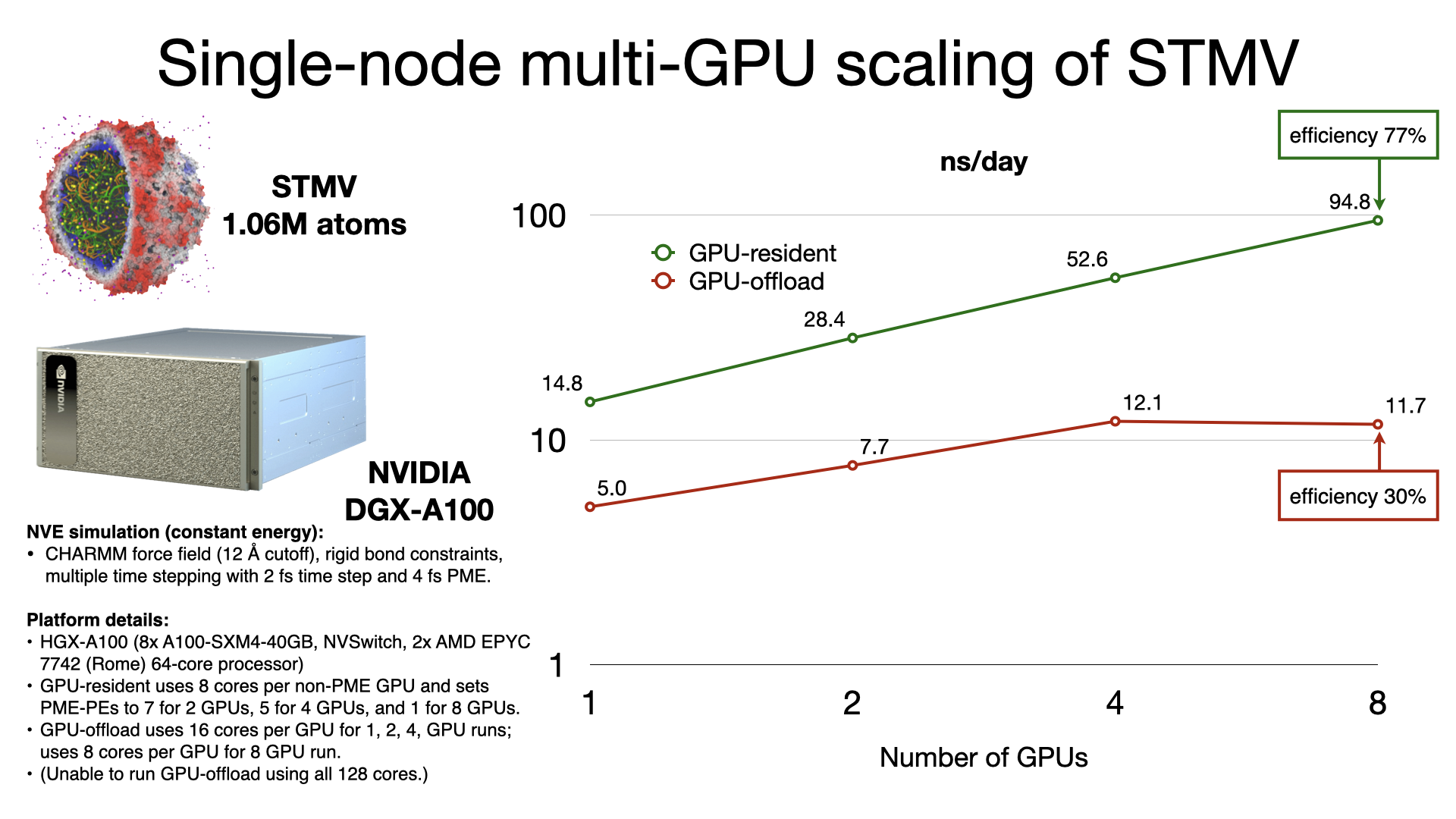
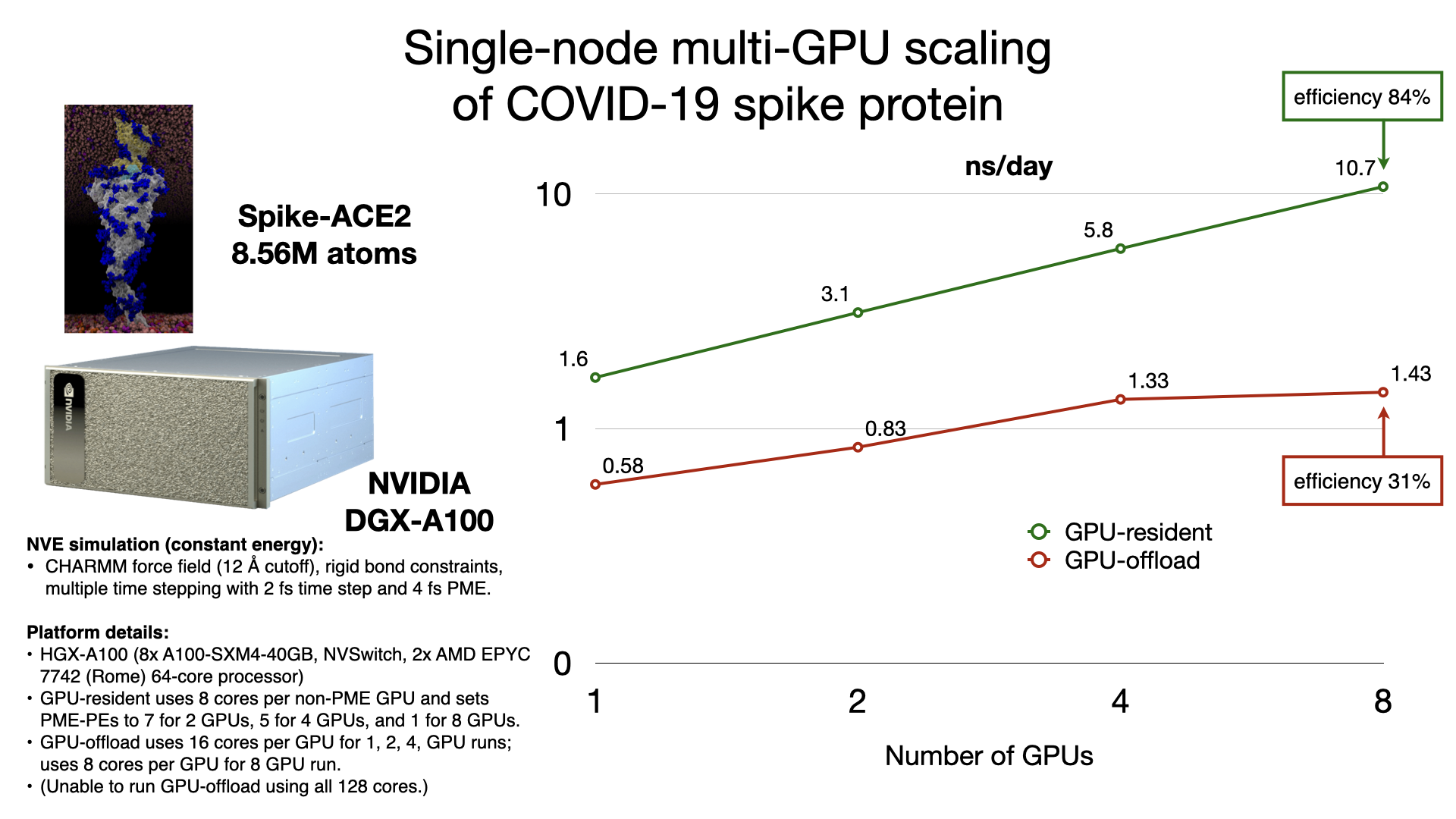
Supported hardware:
GPU-resident and GPU-offload modes are fully supported on NVIDIA GPUs using CUDA, with NVLink or NVSwitch needed for performant multi-GPU scaling. Both modes are also supported on compatible AMD GPUs using HIP, via Hipify translation of the CUDA kernels, with Infinity Fabric needed for performant multi-GPU scaling.
Support for Intel GPUs is still under development, being accomplished by porting NAMD's CUDA kernels to SYCL, utilizing the Intel oneAPI compilers, libraries, and development tools. A 2.15 alpha software release of NAMD contains initial SYCL support for GPU-offload mode for compatible Intel GPUs.
Benchmark files with configuration optimized for GPU-resident simulation mode:
- DHFR (23,558 atoms): dhfr_gpu.tar.gz (1.4M)
- ApoA1 (92,224 atoms): apoa1_gpu.tar.gz (2.8M)
- STMV (1,066,628 atoms): stmv_gpu.tar.gz ( 54M)
NAMD 3.0 parallel scaling results
Benchmarking STMV matrix systems (NPT, 2fs timestep, 12A cutoff + PME every 3 steps) on TACC Frontera
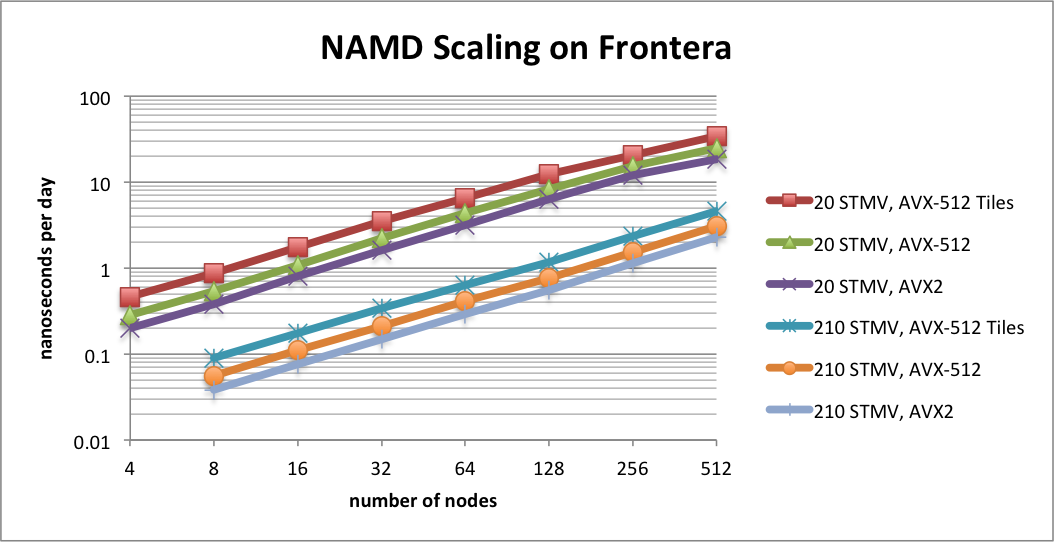
The performance results in this plot show scaling on TACC Frontera for two synthetic benchmark systems comprised of tiling copies of the periodic STMV (satellite tobacco mosaic virus) cell to form arrays of 5x2x2 (21M atoms) and 7x6x5 (224M atoms). The simulations use Langevin piston for pressure control and Langevin damping for temperature control, with a 2fs time step, 12A cutoff, and PME every 3 steps. Special support has been developed in NAMD for CPUs that support AVX-512F instructions, such as the Intel Cascade Lake processors of Frontera and the forthcoming AMD CPUs based on the Zen4 architecture. NAMD 3.0 implements an AVX-512 tile list kernel that is able to achieve up to 1.8x speedup on Intel Xeon processors. The plot compares the new AVX-512 tile list kernel ("AVX-512 Tiles") to the Sky Lake optimized build (Linux-SKX-icc, "AVX-512") and the earlier AVX2 auto-vectorization build (Linux-x86_64-icc, "AVX2").
Benchmarking STMV matrix systems (NPT, 2fs timestep, 12A cutoff + PME every 3 steps) on Summit and Frontera
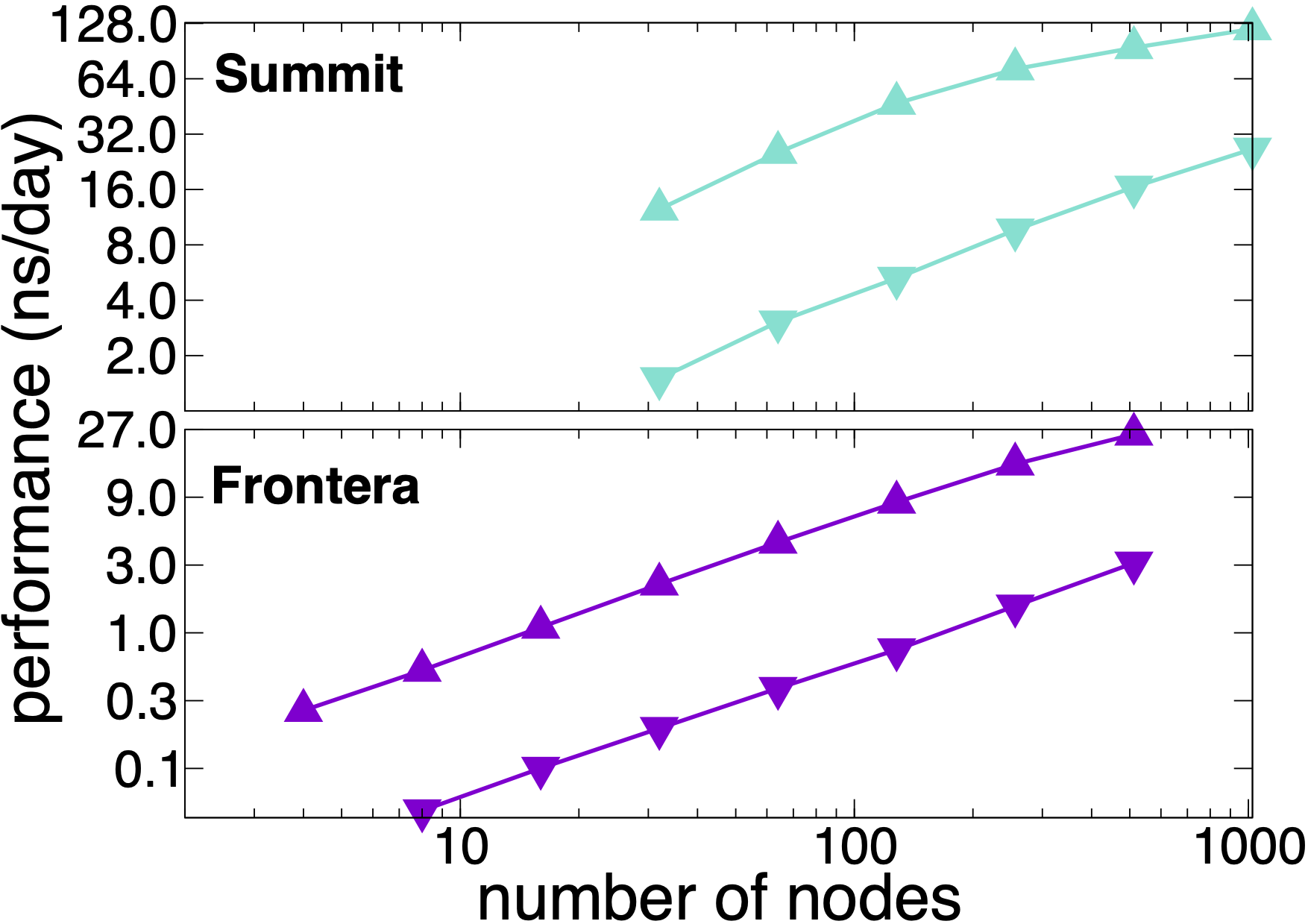
NAMD 3.0 still supports the multi-node scaling available in release 2.14. The plots show performance results on OLCF Summit and TACC Frontera scaling the synthetic benchmark systems created by tiling the periodic STMV (1M atoms) cell in arrays of 5x2x2 (21M atoms, upward pointing triangles) and 7x6x5 (224M atoms, downward pointing triangles). The simulations use Langevin piston and Langevin damping to impose pressure and temperature control (NPT) with a 2fs time step, 12A cutoff, and PME every 3 steps. Multi-node scaling for GPU-based computers like Summit is supported by NAMD's GPU-offload mode.
- STMV (1,066,628 atoms): stmv_gpu.tar.gz ( 54M)
- 20STMV and 210STMV matrix construction: stmv_sc14.tar.gz (2.5K)



