VMD-L Mailing List
From: Nicolas CHARVIN (Nicolas.Charvin_at_univ-savoie.fr)
Date: Wed Apr 06 2005 - 11:10:12 CDT
- Next message: John Stone: "Re: Maximum numbers of atoms"
- Previous message: John Stone: "Re: solvate numbering problems"
- In reply to: John Stone: "Re: Maximum numbers of atoms"
- Next in thread: John Stone: "Re: Maximum numbers of atoms"
- Reply: John Stone: "Re: Maximum numbers of atoms"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]
>Try using the other .vmd files, virus-small.vmd and then virus-large.vmd
>and see if those work on your machine. Let me know if you get the MSMS
>surface as it should be.
after installing MSMS (sorry :)... MSMS 2.5.5 by the way:
virus-simple.vmd loads fine with VDW by default, but when I choose MSMS, I
get many "WARNING: geodesic which intersects odd number of edges" and
"ERROR: find_SES_contact_faces: inf==0 for face 362942", and so on.
Finally, vmd says "failed to compute level 0 surface, try again 2"
virus.vmd (there is no virus-large.vmd on your website) loads, with many
"WARNING: geodesic which intersects odd number of edges" during the MSMS
generation. But it works. I cannot tell you the FPS since it's white
background, but it is approx. 1 fps, and 646 MB of RAM usage.
virus-faster.vmd loads, with "WARNING: Genus mismatch" messages. And it
rotates more smoothly, approx 2 fps, and 413 MB of RAM usage.
Concerning "BigConfig.pdb"
>The "too many bonds" warning indicates that the automatic bond search
>is finding a LOT of atoms spaced so closely together that they are
>considered bonded, the warning indicates that it has gone far past
>the point of any realistic biological structure
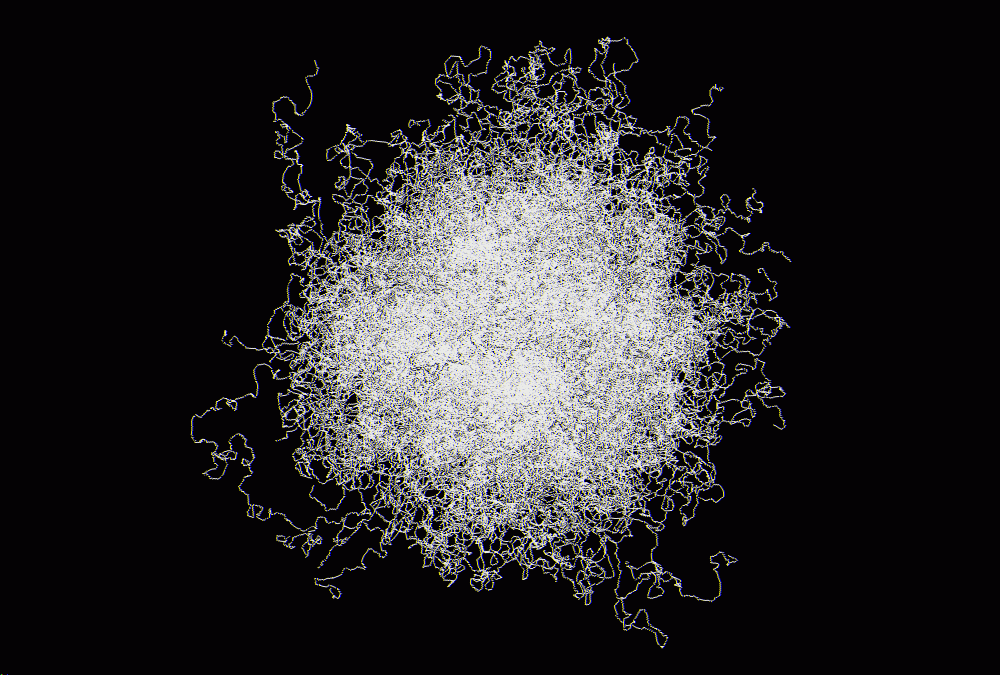
right! It is no biological thing, it is only 640 * 1000 CH2. Please do not
ask me more details ;). I've attached a picture of this molecule under
Qmol (BigConfig_under_Qmol.gif).
Thanks a lot for your help
nicolas
-- Nicolas.Charvin_at_univ-savoie.fr LMOPS - Lab. Matériaux Organiques à Propriétés Spécifiques Bât IUT - Université de Savoie 73376 LE BOURGET DU LAC CEDEX Tel: 04-79-75-86-53 http://www.univ-savoie.fr/labos/lmops
- Next message: John Stone: "Re: Maximum numbers of atoms"
- Previous message: John Stone: "Re: solvate numbering problems"
- In reply to: John Stone: "Re: Maximum numbers of atoms"
- Next in thread: John Stone: "Re: Maximum numbers of atoms"
- Reply: John Stone: "Re: Maximum numbers of atoms"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]



