VMD-L Mailing List
From: Luis Rosales (ludwig_at_biomedicas.unam.mx)
Date: Tue Oct 26 2004 - 15:11:53 CDT
- Next message: Cheri M Turman: "Re: error while loading shared libraries: libGLU.so.1"
- Previous message: John Stone: "Re: Problems with povray"
- In reply to: John Stone: "Re: Problems with povray"
- Next in thread: John Stone: "Re: Problems with povray"
- Reply: John Stone: "Re: Problems with povray"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]
Dear John,
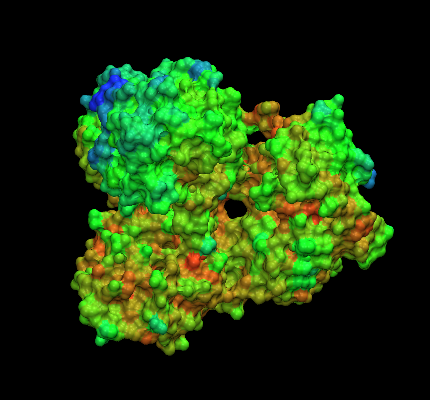
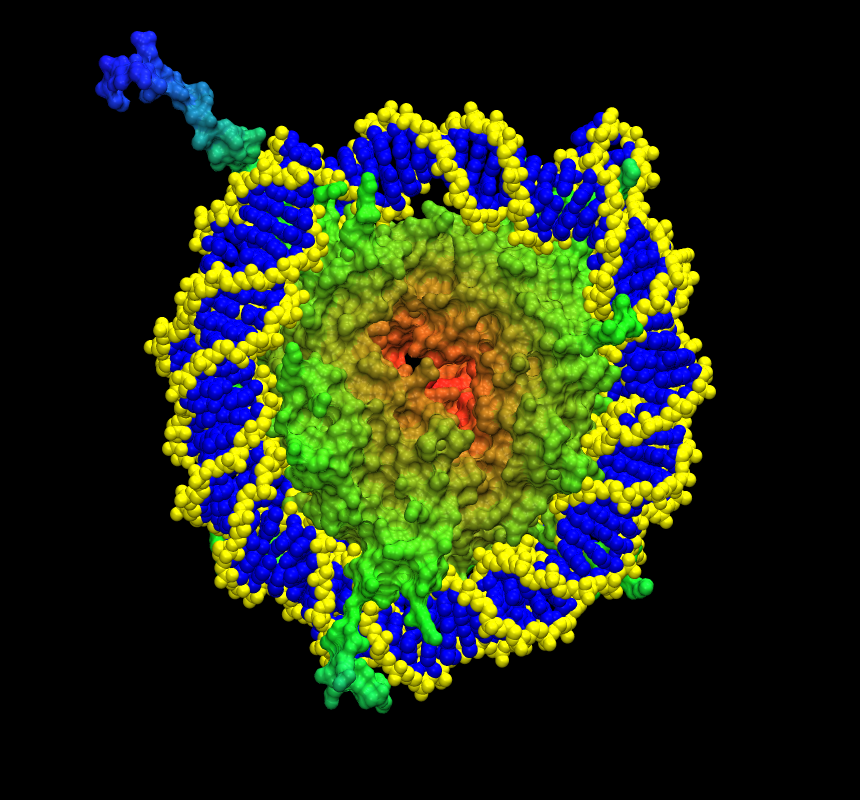
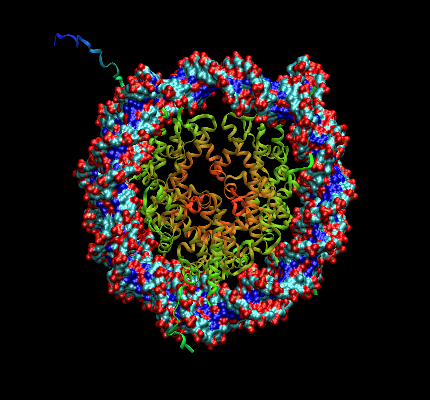
I just tried the new alpha version of vmd (Linux)!!! (a couple of images are
attached on the mail...)
The Pov export of surfaces is really good, a lot cleaner than the previous
versions.
I generated surfaces on small proteins like ubiquitin and big complexes like
the nucleosome, on every case the parsing of the files was just fine,
without the degenerate triangle warning.
Also, I think that the size of the generated Pov files is a lot smaller than
the previous versions (I only checked ubiquitin and the size difference was
8Mb in vmd 1.8.3a vs. 11Mb in vmd 1.8.2).
The only trouble I found was on the rendering of the "new ribbons"
representation of proteins. On every case I got the message "all
determinants too small". Even as I tried warious ribbons resolutions, I was
unable to avoid the warning. I am not sure, but I wonder if this could be
related to the curvature asociated to the "sides" of the ribbon??
One last question,
I wonder why color gradients are not conserved when you render them on a
surface, (why a surface colored by position is made of discrete colored
"patches" of tringles instead of a true gradient?).
Thank you and best regards,
Luis
> -----Mensaje original-----
> > >
> > > I tested on both linux and windows (I use VMD 1.8.2 and Povray 3.5),
> > > but to no avail. I used VMD/povray already before, and I don't
remember
> > > to have had problems then. My structures consists mainly of
> > > Bonds, VDW and NewRibbons Graphical Representations.
> > >
> > > Any clues?
> > >
> > > Regards,
> > >
> > > Dominique
- Next message: Cheri M Turman: "Re: error while loading shared libraries: libGLU.so.1"
- Previous message: John Stone: "Re: Problems with povray"
- In reply to: John Stone: "Re: Problems with povray"
- Next in thread: John Stone: "Re: Problems with povray"
- Reply: John Stone: "Re: Problems with povray"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]



