VMD-L Mailing List
From: Bennion, Brian (Bennion1_at_llnl.gov)
Date: Thu Sep 18 2014 - 17:21:43 CDT
- Next message: John Stone: "Re: Extension crashes"
- Previous message: Ana Celia Vila Verde: "Re: measure hbonds in VMD tcl script"
- Next in thread: Tristan Croll: "Re: using vmd to apply symmetry operators part 2"
- Reply: Tristan Croll: "Re: using vmd to apply symmetry operators part 2"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]
Hello,
I posted a similar question a few years back and was directed to the mono2poly tcl script.
http://www.ks.uiuc.edu/Research/vmd/mailing_list/vmd-l/19379.html
I would like to add onto this discussion as recent work forced my to look at this again.
Using the mono2Poly script appears to work as written for expanding out the structure of the asymmetric unit to the biologically relevant assembly.
Now, if i wanted to recreate the unit cell I should be able to use the same type of matrix operations with the symtry records in the pdb file as found in the remarks 290 section. The unit cell has dimensions 33.95 33.95 74.82.
Initially I just modified the mono2poly.tcl to search for the symtry string to populate the matrices. In my case I was using 2FDN which has a single chain in the asymmetric unit but is a 8mer in the unit cell. So there are 8 symtry operations found by mono2poly.
The resulting structure of the unit cell is wrong.
So I try it by hand and entered a few of the symtry operations into matrices sym[1-8] and used the $all move $sym[1-8] commands to update the coordinates.
example:
REMARK 290 SMTRY1 8 0.000000 -1.000000 0.000000 0.00000
REMARK 290 SMTRY2 8 -1.000000 0.000000 0.000000 0.00000
REMARK 290 SMTRY3 8 0.000000 0.000000 -1.000000 37.41000
set sym8 {{0.0 -1.0 0.0 0.0 } { -1.0 0.0 0.0 0.0} { 0.0 0.0 -1.0 37.41 } { 0.0 0.0 0.0 1.0 }}
I was only able to replicate what the mono2poly script had done previously.
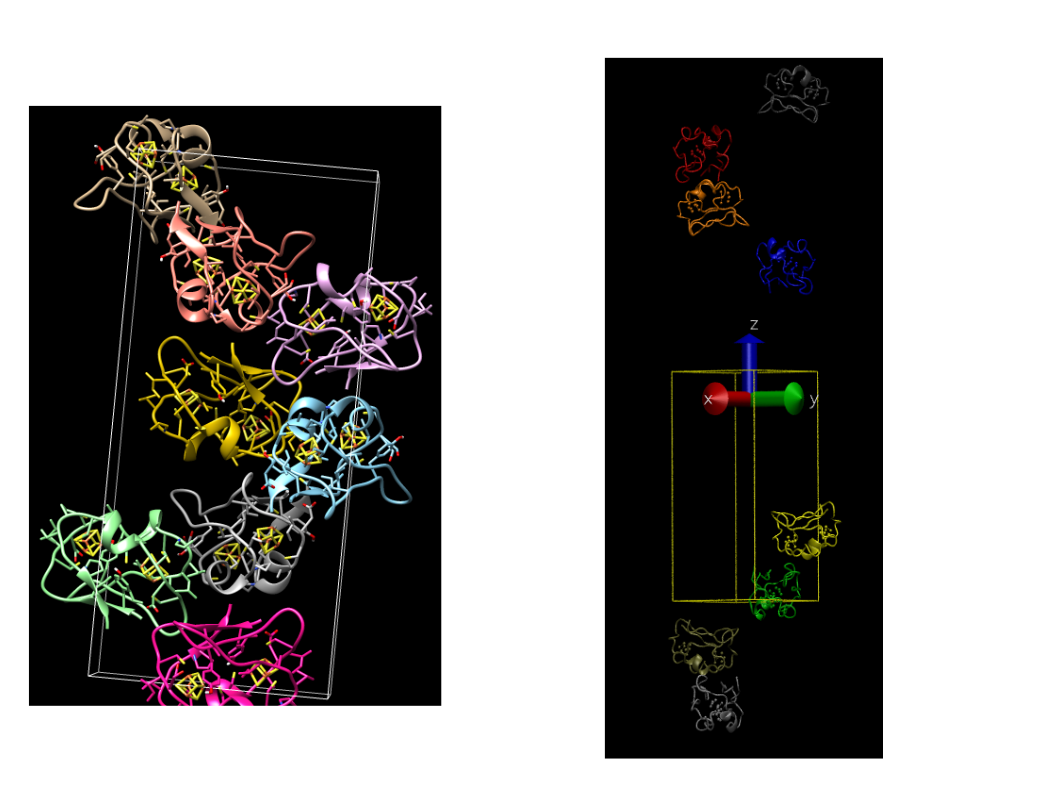
Attached is an image that compares what chimera outputs and what the mono2poly script outputs.
Did I create the matrices incorrectly?
Brian
- Next message: John Stone: "Re: Extension crashes"
- Previous message: Ana Celia Vila Verde: "Re: measure hbonds in VMD tcl script"
- Next in thread: Tristan Croll: "Re: using vmd to apply symmetry operators part 2"
- Reply: Tristan Croll: "Re: using vmd to apply symmetry operators part 2"
- Messages sorted by: [ date ] [ thread ] [ subject ] [ author ] [ attachment ]



