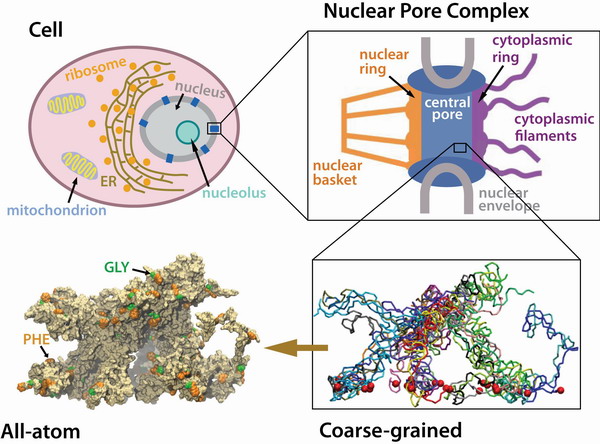
The Nuclear Pore Complex
Gateway to the Nucleus
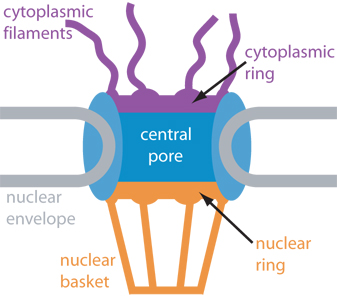 Schematic cross-section of
the nuclear pore complex showing major components
Schematic cross-section of
the nuclear pore complex showing major components
The nucleus of the cell is centrally important to an organism. It serves to store and organize genetic information, the atomic blueprint for the organism, while separating and protecting this very important information from the host of other cellular components. While the nucleus requires this protective isolation, it also needs to communicate with the rest of the cell, exchanging proteins and RNA, for a variety of nuclear and cytoplasmic processes which act in concert. The nuclear pore complex (NPC), perhaps the largest protein complex in the cell, is responsible for the protected exchange of components between the nucleus and cytoplasm and for preventing the transport of material not destined to cross the nuclear envelope.
Much is known about the behemoth complex from a variety
of experimental techniques, but its vast size has made
pointed study quite difficult. The NPC has a mass of ~125
megaDaltons
(MDa) in vertebrates
(![]() )
and 66 MDa in yeast (
)
and 66 MDa in yeast (![]() ).
It has an octagonally symmetric
cylindrical structure around the axis of transport and a planar
pseudo-symmetry through the nuclear envelope. The yeast NPC has
been well characterized and is thought to contain an upper limit of
30 distinct types of proteins (termed nucleoporins, or nups)
(
).
It has an octagonally symmetric
cylindrical structure around the axis of transport and a planar
pseudo-symmetry through the nuclear envelope. The yeast NPC has
been well characterized and is thought to contain an upper limit of
30 distinct types of proteins (termed nucleoporins, or nups)
(![]() ).
The number is low and rather surprising considering that
the
ribosome,
another macromolecular protein
complex, contains 75 different types of proteins and weighs only
4 MDa. The body of the vertebrate NPC is approximately 145 nm in
diameter and 80 nm length across the nuclear envelope. The yeast NPC
is smaller at approximately 96 nm in diameter and 35 nm in length
(
).
The number is low and rather surprising considering that
the
ribosome,
another macromolecular protein
complex, contains 75 different types of proteins and weighs only
4 MDa. The body of the vertebrate NPC is approximately 145 nm in
diameter and 80 nm length across the nuclear envelope. The yeast NPC
is smaller at approximately 96 nm in diameter and 35 nm in length
(![]() ).
The NPC is truly large, especially when compared to other proteins
or protein complexes in the cell. For comparison, a tetramer of aquaporin proteins measures only 6.5 nm
across!
).
The NPC is truly large, especially when compared to other proteins
or protein complexes in the cell. For comparison, a tetramer of aquaporin proteins measures only 6.5 nm
across!
Nucleocytoplasmic Transport
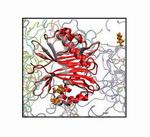
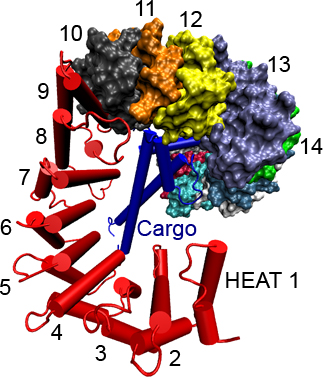 Importin-β (red) with
cargo SREBP-2 fragment (blue) bound. Dual α-helical repeats
(HEAT repeats), which comprise the structure of importin-β,
are numbered.
Importin-β (red) with
cargo SREBP-2 fragment (blue) bound. Dual α-helical repeats
(HEAT repeats), which comprise the structure of importin-β,
are numbered.
The immense size of the NPC makes it difficult to study the system in detail both experimentally and computationally. However, a variety of experiments have elucidated many properties of the NPC and the transport process itself. They have described the macroscopic structure of the NPC as well as other features which provide the foundation for computational efforts to tackle the mechanism of nucleocytoplasmic transport.
Transport Cycle.
In order to pass through the NPC, a large molecule (cargo) must
associate with another protein called a transport receptor. These
transport receptors then act as chaperones which shuttle cargo
through the NPC. Recognition by the transport receptor takes place
via a specific sequence of amino acids in the cargo protein. Besides
cargo and transport receptors, the third ingredient necessary for
nuclear transport is the signaling protein
Ran,
which hydrolyzes
GTP.
Ran is responsible for regulating the interaction of transport
receptor and cargo
(![]() ,
,
![]() ),
and RanGDP/RanGTP concentration gradients across the nuclear envelope
drive nuclear import and export. Once a transport receptor identifies and
binds its cargo, the transport complex may be imported or exported through
the NPC. The mechanism by which the NPC selectively allows the transit of
import or export complexes, while restricting the passage of inert species
is poorly understood. However, nups with repeating amino acid sequences
involving phenyalanine (F) and glycine (G) have been thoroughly implicated
in the process.
),
and RanGDP/RanGTP concentration gradients across the nuclear envelope
drive nuclear import and export. Once a transport receptor identifies and
binds its cargo, the transport complex may be imported or exported through
the NPC. The mechanism by which the NPC selectively allows the transit of
import or export complexes, while restricting the passage of inert species
is poorly understood. However, nups with repeating amino acid sequences
involving phenyalanine (F) and glycine (G) have been thoroughly implicated
in the process.
FG-Nups.
The NPC requires the presence of nups with FG sequence
repeats. Many transport receptors, such as importin-β
(![]() ,
,
![]() ,
,
![]() )
and NTF2
(
)
and NTF2
(![]() ,
,
![]() ,
,
![]() ,
,
![]() ),
have been shown to have hydrophobic
binding sites on their surface for these "FG-Nups". Furthermore,
the systematic deletion of the FG-repeat regions from FG-Nups
in yeast NPCs has been shown to be lethal to yeast cells
(
),
have been shown to have hydrophobic
binding sites on their surface for these "FG-Nups". Furthermore,
the systematic deletion of the FG-repeat regions from FG-Nups
in yeast NPCs has been shown to be lethal to yeast cells
(![]() ).
The deletion of the
FG-repeat region from one nup, nup116p, caused cell death on
it own. FG-Nups have also been shown to form filaments
(
).
The deletion of the
FG-repeat region from one nup, nup116p, caused cell death on
it own. FG-Nups have also been shown to form filaments
(![]() )
and colocalize with the filaments on both sides of the NPC. This evidence
is consistent with the FG-Nups forming the filaments of the NPC, to
which transport complexes may dock in the initial and final stages of
transport. Finally, the FG-Nups have been shown to be highly disordered
and contain almost no secondary structure
(
)
and colocalize with the filaments on both sides of the NPC. This evidence
is consistent with the FG-Nups forming the filaments of the NPC, to
which transport complexes may dock in the initial and final stages of
transport. Finally, the FG-Nups have been shown to be highly disordered
and contain almost no secondary structure
(![]() ).
Localization of nups in the yeast NPC
(
).
Localization of nups in the yeast NPC
(![]() )
shows that many of the FG-Nups occupy the central pore region of the NPC,
through which the transport complexes pass. All this evidence points
toward intimate interactions which must take place between FG-Nups and
transport receptors to make transport successful.
)
shows that many of the FG-Nups occupy the central pore region of the NPC,
through which the transport complexes pass. All this evidence points
toward intimate interactions which must take place between FG-Nups and
transport receptors to make transport successful.
The Selective Barrier.
The gating mechanism of NPC selective transport remains elusive. Due to the disordered nature of FG-nups, a detailed
understanding of the selective barrier is hard to be achived.
Various models,
such as the virtual-gate
(![]() ),
the selective phase
(
),
the selective phase
(![]() ),
and the reduction-of-dimensionality
(
),
and the reduction-of-dimensionality
(![]() )
model,
have been proposed to explain the NPC selective barrier and thus the transport. While FG-nups are
beleived to form the selective barrier, the exact form of the barrier is still unkown.
)
model,
have been proposed to explain the NPC selective barrier and thus the transport. While FG-nups are
beleived to form the selective barrier, the exact form of the barrier is still unkown.
Click the picture below for
a movie (mpeg, [an error occurred while processing this directive]) showing
peptides of amino acid sequence GLFG binding to the surface of
importin-β.

Discovering New Transport Receptor Interactions
In order to understand the mechanism of nucleocytoplasmic transport, one must understand the manner in which the NPC interacts with transport receptors. To examine these interactions, we have performed simulations of transport receptors in a solution containing short chains of FG-Nups. We have examined both the transport receptors importin-β and NTF2. During the course of any single simulation, FG-Nups were found to diffuse through the solution and bind to the surface of the transport receptor. Some of these interactions are likely to play a role in the in vivo recognition of the transport receptor by the NPC, and allowing it passage into or out of the nucleus.
To determine which of the several binding events occurring in a simulation are relevant, we also aligned several sequences of the transport receptor, with each set of aligned residues receiving a score based on conservation, evolutionary distance (see our tutorial on sequence alignment [pdf]), and hydrophobicity. Highly scoring residues were regarded as "hot spots" for the binding of FG-Nups on the surface of the transport receptor (see importin-β figure on the right).
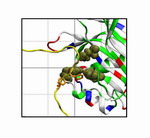
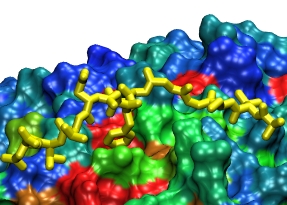 FG-Nup (yellow) binding to the
surface of importin-β at a conserved binding "hot spot" (red)
between HEAT repeats 6/7
FG-Nup (yellow) binding to the
surface of importin-β at a conserved binding "hot spot" (red)
between HEAT repeats 6/7
Importin-β.
We simulated a crystal structure
(![]() )
of the transport receptor importin-β with cargo, fully solvated and
ionized, in the presence of FG-Nups, and performed a sequence alignment using
eight importin-β species. Using this combination of molecular dynamics
simulations and sequence alignment, we were able to confirm the existence
of 3 out of 4 binding spots which were known previously from experiments
(
)
of the transport receptor importin-β with cargo, fully solvated and
ionized, in the presence of FG-Nups, and performed a sequence alignment using
eight importin-β species. Using this combination of molecular dynamics
simulations and sequence alignment, we were able to confirm the existence
of 3 out of 4 binding spots which were known previously from experiments
(![]() ,
,
![]() ,
,
![]() ).
Furthermore, we were able to identify six novel binding spots for FG-Nups
which were previously unknown. (One of these spots was identified
independently by experimentalists
(
).
Furthermore, we were able to identify six novel binding spots for FG-Nups
which were previously unknown. (One of these spots was identified
independently by experimentalists
(![]() ).)
The simulations provide the first atomic-scale look at these novel binding
spots for FG-Nups, and suggest that the extent of binding on the surface
of importin-β is much larger than previously realized. The additional
binding spots on the surface of importin-β suggest that the molecule
has a more extensive ability to interact with the NPC than previously
expected and also implies that other transport receptors similar in size
harbor an equally large number of binding spots.
).)
The simulations provide the first atomic-scale look at these novel binding
spots for FG-Nups, and suggest that the extent of binding on the surface
of importin-β is much larger than previously realized. The additional
binding spots on the surface of importin-β suggest that the molecule
has a more extensive ability to interact with the NPC than previously
expected and also implies that other transport receptors similar in size
harbor an equally large number of binding spots.
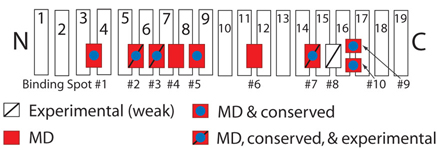
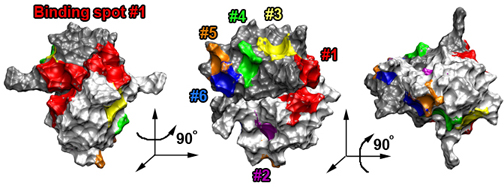
Schematic picture of importin-β showing binding spots for FG-Nups on its surface.
Those spots discovered experimentally are labeled with a slash, while those uncovered in our simulations are red.
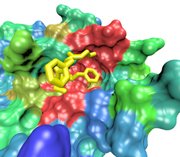 FG-Nup (yellow) binding to NTF2
at the spot where Ran binds upon import
FG-Nup (yellow) binding to NTF2
at the spot where Ran binds upon import
NTF2. The transport receptor NTF2 is a vital in maintaining the proper balance of components across the nuclear envelope to drive nuclear import and export. The protein Ran is an enzyme which catalyzes the hydrolysis of GTP. The energy released upon hydrolysis is used to disassemble transport complexes which have been exported to the cytoplasm. Alternatively, the binding of RanGTP to transport receptors may initiate transport complex formation (for those to be exported) or initiate disassembly (for those which have been imported). Maintaining the proper balance of Ran across the nuclear envelope is crucial for nuclear transport. The reimporting of RanGDP to the nucleus, where it is charged to become RanGTP, is the job of NTF2.
We performed two sets of simulations on NTF2. One set
involved NTF2 with RanGDP bound
(![]() ),
as the transport complex would appear
upon import into the nucleus. The other set involved the simulation of
NTF2 alone
(
),
as the transport complex would appear
upon import into the nucleus. The other set involved the simulation of
NTF2 alone
(![]() ),
as the dimer would appear upon export. We simulated both
complexes in a solution of short chains of FG-Nups for a total of over 250 ns
to determine how and where the chains interacted with the NTF2 surface, and
aligned the sequences of 15 distinct NTF2 species to support the simulation
findings. Using the technique, we were able to verify the existence of four
binding spots on the NTF2 surface already discovered via previous
X-ray (
),
as the dimer would appear upon export. We simulated both
complexes in a solution of short chains of FG-Nups for a total of over 250 ns
to determine how and where the chains interacted with the NTF2 surface, and
aligned the sequences of 15 distinct NTF2 species to support the simulation
findings. Using the technique, we were able to verify the existence of four
binding spots on the NTF2 surface already discovered via previous
X-ray (![]() ),
mutational (
),
mutational (![]() ,
,
![]() ,
,
![]() ),
NMR (
),
NMR (![]() ),
and computational data (
),
and computational data (![]() ).
We also discovered two novel
binding spots, not yet discovered by experiment. All six binding spots
broadly form a stripe across the surface of NTF2, as indicated in the
figure below. The regularity and proximity of binding spots play a role
in the identification of the NTF2 by NPC FG-Nups. Furthermore, the novel
binding spots appear on a structurally variable part of the NTF2 surface,
suggesting that these spots, along with the Ran binding spot, may play a
role in tuning the transport of NTF2 for import or export.
).
We also discovered two novel
binding spots, not yet discovered by experiment. All six binding spots
broadly form a stripe across the surface of NTF2, as indicated in the
figure below. The regularity and proximity of binding spots play a role
in the identification of the NTF2 by NPC FG-Nups. Furthermore, the novel
binding spots appear on a structurally variable part of the NTF2 surface,
suggesting that these spots, along with the Ran binding spot, may play a
role in tuning the transport of NTF2 for import or export.
Schematic picture of NTF2 showing the six binding spots for FG-Nups on its surface.
Each NTF2 dimer is shown in a different shad of grey.
The existence of numerous binding spots on the surface of any transport receptor can serve to distinguish it from other proteins which reside around the nuclear envelope or it may enable the NPC to more finely tune the transport of cargo. Upon examination of a third transport receptor, Cse1p, the case becomes convincing for numerous binding spots on the transport receptor surface as a necessity for nuclear transport.
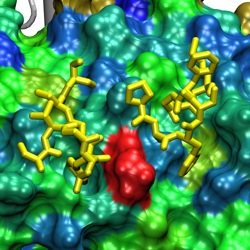 Two FG-Nups (yellow) binding to the
surface of Cse1p at a conserved binding spot (red)
between HEAT repeats 12/13/14
Two FG-Nups (yellow) binding to the
surface of Cse1p at a conserved binding spot (red)
between HEAT repeats 12/13/14
Cse1p.
Cse1p is a yeast transport receptor which functions as the Kap60p exporter.
(Cse1p is known as CAS in vertebrates and Kap60p is importin-α)
(![]() ,
,
![]() ,
,
![]() ,
,
![]() ).
The transport receptor recycles Kap60p back to the cytoplasm after it assists
in nuclear import
as an adaptor for Kap95p (importin-β). The crystal structure of Cse1p is
available in complex with Kap60p and RanGTP
(
).
The transport receptor recycles Kap60p back to the cytoplasm after it assists
in nuclear import
as an adaptor for Kap95p (importin-β). The crystal structure of Cse1p is
available in complex with Kap60p and RanGTP
(![]() )
and in cargo-free form
(
)
and in cargo-free form
(![]() ).
Like importin-β, Cse1p is a
right-handed superhelical protein. It is composed of 20 HEAT repeats.
Kap60p is comprised primarily of 10 Armadillo (ARM) repeats.
).
Like importin-β, Cse1p is a
right-handed superhelical protein. It is composed of 20 HEAT repeats.
Kap60p is comprised primarily of 10 Armadillo (ARM) repeats.
We performed four molecular dynamics simulations on the Cse1p:Kap60p:RanGTP export complex, similar to those performed on importin-β and NTF2, to study interactions between FG-nup peptides and the surface of the transport complex. The simulations revealed 14 binding spots for FG-repeat peptides on the surface of Cse1p and five binding spots on the surface of Kap60p. They are particularly valuable in this case because little study has been devoted to detailing the interactions of FG-Nups with Cse1p compared to the large body of evidence uncovered for importin-β and NTF2. We found binding of FG-Nups to be concentrated between HEAT repeats 10-16, with eight of the 14 binding spots appearing in that region and one or more binding spots for each crevice between repeats. Binding of FG-nup peptides to Kap60p, on the other hand, appears to be much less extensive and sparser than on the Cse1p surface at only five spots on the Kap60p surface, indicating a much lesser concentration in binding spots than seen on the Cse1p surface. The similarities in FG-Nup binding between Cse1p and the transport receptors importin-β and NTF2 and the differences in binding of Kap60p, an inert macromolecule that cannot traverse the nuclear pore alone, provide a unique insight into how the nuclear pore likely recognizes a transport receptor (and cargo) destined for nuclear import, while preventing the transport of other molecules.
Comparing the binding spots on the surfaces of the three transport receptors with that of Kap60p, reveals a striking difference, not simply in the number of binding spots, but most notably in their orientation on the surfaces of each protein. The relatively small amount of Kap60p binding spots yields a lower average binding spot density across the entire protein surface than across that of Cse1p, for example. Furthermore, the Kap60p binding spots are also spread widely across the protein's surface from ARM repeats 1-9, with no more than two binding spots clustered at short distances. The location of Cse1p binding spots, on the other hand, appears in three broader clusters across the surface, the most populated one near the beginning of the C-terminal half of the molecule: eight of the 14 Cse1p binding spots are located in this cluster between HEAT repeats 10-16. The binding spots in this cluster are separated from their neighboring binding spots by regular distances of ~16 Å (± 4 Å). Importin-β and NTF2 exhibit similar sets of concentrated binding spots on their surfaces.
The multitude of binding spots separated by regular and short distances on the surfaces of the Cse1p, importin-β, and NTF2 compared to that of Kap60p suggests that the NPC uses both binding spot number and proximity to determine whether a macromolecule (complex) is fit for transport. In distinguishing between legitimate transport receptors and inert molecules not destined to traverse the nuclear envelope alone, the NPC must utilize a mechanism which is capable of protecting against the random occurrence of a binding spot on the surface of an inert molecule that is capable of binding FG-nups. Indeed, any inert macromolecule may possess one or several hydrophobic depressions on its surface not explicitly for nuclear transport but which are capable of FG-nup binding nonetheless. The binding of a single FG-repeat, or even a small few, to the surface of a molecule is unlikely to render successful nuclear transport. The NPC may protect against these "random" binding spots by requiring a transport receptor to have several binding spots in close proximity on its surface. With numerous and close binding spots on a transport receptor's surface, the NPC may be able to bind multiple FG-repeats to the transport receptor simultaneously, ensuring a strong enough composite binding energy to enable nuclear transport. Furthermore, the regular spacing between binding spots may be coordinated with the regular spacing between FG- repeats within individual FG-nups. Both spacings may be tuned to allow the binding of FG-repeats to binding spots, while also allowing the energetically favorable adherence of FG-nup linker regions to the transport receptor surface. This regular and numerous binding of FG-nups may effectively serve as a robust means for the NPC to identify transport complexes among a host of inert molecules that may possess hydrophobic patches on their surfaces.
Click the picture below for
a movie (mpeg, [an error occurred while processing this directive]) showing
the location of FG-Nup binding spots on the sufaces of Cse1p and
Kap60p.

Exploring the Structure of the Selective Barrier.
The NPC controls passage into and out of the nucleus through an amazing filter. Like an ordinary filter the NPC permits passage of small particles (biomolecules), but not of large particles (e.g., proteins). However, certain large particles, namely the transport receptors, can pass the NPC. For this purpose the NPC employes long "finger" proteins anchored on the NPC perimeter. The transport receptors apparently can intermittently widen the filter pores while other proteins cannot. To observe how this is achieved is difficult since the finger proteins are highly disordered. Molecular dynamics simulations may offer an answer, though, as they have been very successful in describing in structural and dynamic detail another disordered cellular system, lipid membranes. Accordingly, we have applied the molecular dynamics approach to study the transport-related structures and processes inside the NPC through the same methods earlier applied to lipid membrane processes, namely coarse-grained and atom level simulations. The key question we tried to answer was: what kind of structure do assemblies of finger proteins form and how do they relate to selective transport in the NPC.
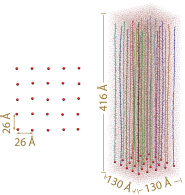
The initial array. Left: top view of the array. Only tetherring points are shown. Right: side view of the array. Each segment is depicted in a different color. Neighboring periodic cells are not shown.
In our simulations we brought finger proteins together, namely FG-nups. These proteins are characterized through FG repeats. For isolated FG-nups we found that the proteins coil-up into a globular shape with most FG repeats hidden inside the globule to protect these hydrophobic groups from water. We then brought the FG-nups together at a density that corresponds to the FG-repeat density in the NPC central channel. For this purpose we tethered an array of stretched-out FG-nups (wild-type and mutant yeast nsp1) segments to a planar surface as shown in the figure and the movie. By combing coarse-grained (CG) and all-atom (AA) simulations, we were able to study over a time scale of microseconds how the stretched-out and initially separated FG-nups formed rapidly bundles of 2-6 proteins while collapsing into a brush-like system shown below. The bristles of the brush are bundles of FG-nups and have two key properties as shown below: (i) on their surface they are dotted with spots of FG pairs, that interact favorably with transport receptors (see above); (ii) the bristles are also interconnected, namely where finger proteins change from one bundle to another bundle, which they do with some frequency (see the figures and movies below). It appears then that the bristles of the nuclear pore filter form an energetically favorable environment (through their many FG-spots) for transport receptors and that the latter can tear a finger protein readily away from a bundle to form a wider space for passage. While segments individually simulated were seen to form globule-like dynamic structures, arrays of them formed bundle-based brush-like structures with a height much larger than the Rg of individually simulated segments. All-atom simulations were also performed to investigate the interaction between the brush-like structure and the transport receptor NTF2.

An array of stretched out FG-nups coiled and shortened into a dynamic bundle-based brush-like structure. Movie 1 (mpg 8.7M) shows the formation process in more detail. Movie 2 (mpg 4.6M) and movie 3 (mpg 4.7M) show further detail of the resulting brush-like structure.
For our simulations the FG-repeat domain of nsp1 (1-600) was divided into 25 100 amino acid (aa) long segments (D1-D25, Table). The resulting 25 segments were then attached individually to a planar surface, forming a 5 X 5 array, to sample computationally a representative volume of the nsp1-filled NPC central channel. The tethering points were positioned so that the final, i.e., collapsed, system, adopted an FG-repeat density similar to that in NPC channels. Two 4 microseconds of coarse-grained (CG) simulations were then performed on the array system, each followed by a 10 ns all-atom (AA) simulation. While the CG simulations extend the timescale of our simulations to microseconds, allowing nsp1 segments to self-aggregate, the AA simulations refined the obtained CG structures. For comparison and inspired by experimental work
(![]() ), an array was simulated with mutant FG-nups in which most PHEs of the wild-type were replaced by SERs. This simulation followed the same procedure, i.e., involved a combination of a 4 microsecond CG simulation and a subsequent 10 ns AA simulation. For further comparison, six representative FG-nup segments, also tethered to a planar surface, were studied individually by performing a one microsecond CG MD and subsequently a 30 ns AA simulation.
), an array was simulated with mutant FG-nups in which most PHEs of the wild-type were replaced by SERs. This simulation followed the same procedure, i.e., involved a combination of a 4 microsecond CG simulation and a subsequent 10 ns AA simulation. For further comparison, six representative FG-nup segments, also tethered to a planar surface, were studied individually by performing a one microsecond CG MD and subsequently a 30 ns AA simulation.
In case of individual FG-nups, the initially fully extended segments coiled and shortened very quickly into globule-like structures, with a radius of gyration (Rg) ranging from 13.7 Å to 20.0 Å for the six simulations. For both wild-type and mutant array simulations, the segments collapsed from their fully extended state, reaching a dynamic equilibrium very quickly. But instead of forming globule-like structures, the array in each simulation reached a dynamic bundle-based brush-like structure with a brush height much larger than the Rg value obtained in the individual segment simulations. The wild-type array reached a brush height of 77.1 Å for one simulation and 88.8 Å for the other simulation. The mutant array reached an even higher brush-like structure, with a height of 114.4 Å . Surface representations of both wild-type brush-like structures show many FG-repeats exposed on the brush-surface, offering a favorable environment for transport receptors.
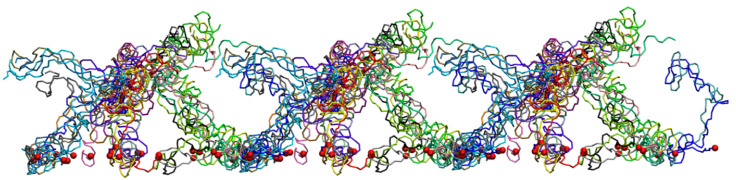
One of the wild-type brush-like structures at the end of its 4 us CG simulation. Neighboring periodic cells are also shown.
Each segment is depicted in a different color and the constrained N-termini (tethering points) are represented by red spheres.
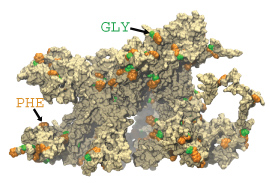 One wild-type brush-like structure at the end of its AA simulation is shown in surface representation.
FG-repeats are shown in green and orange.
One wild-type brush-like structure at the end of its AA simulation is shown in surface representation.
FG-repeats are shown in green and orange.
To further identify how FG-repeats and the mutated SG-repeats were distributed in the final brush-like structures, the pair distribution function g(r) for the FG/SG-repeats was calculated by averaging over the last 2 ns of each array AA simulation. Although the two wild-type simulations reached quite different final conformations, the g(r) for both of them exhibit a similar FG-FG distance distribution at the first peak of g(r) located at r ~ 6Å . Further more, sampling the FG-FG distance with a cutoff of 8 A, the edge of the first peak, revealed that 41.5% of the FG-repeats were involved in close contact in one wild-type simulation and 37.5% in the other. In other words, more than half of the FG-repeats don't have any other FG-repeat neighboring within a distance of 8 Å . These FG-repeats should be free to bind to transport receptors. On the other hand, the g(r) of segments with (mutated) SG-repeats looks quite different from the wild-type ones. Instead of forming a main first peak, the corresponding g(r) is seen to split into three lower peaks. Further calculation showed that 31.1% SG-repeats were in close contact, i.e., less than in the wild-type case. Given that the mutant brush was also ~20 Å higher than the wild-type brushes, the FG-repeats seem to play a role in affecting the overall appearance of the brush-like structure, but apparently they do not dominate the formation of the brushes.
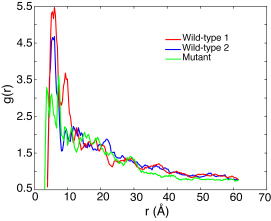 Pair distribution function of the FG/SG-repeats of all three array simulations averaged over the last 2 ns of their AA simulations.
Pair distribution function of the FG/SG-repeats of all three array simulations averaged over the last 2 ns of their AA simulations.
To investigate how the brush-like structures interact with transport receptors, one NTF2 dimer (this transport receptor is also discussed further above) was added into one of the final all-atom wild-type brush-like structures and the system was then simulated for 60 ns in AA MD. Two NTF2 binding spots were seen bound to FG-repeat PHEs, with one FG-repeat even coming from further away. Although the FG-repeat was constrained by the bundle-based brush-like structure, it was still able to come closer and bind to the NTF2 surface. The ability of binding to different FG-repeats of different segments and the ability of engaging further away FG-repeats should be important for NTF2's transport through the NPC.
Two different mechanisms suggested among others for NPC gating, are the selective-phase model
(![]() )
and the virtual-gate model
(
)
and the virtual-gate model
(![]() ). The main difference between these models
is the degree to which FG-FG binding is involved in forming the gating barrier in the NPC. Our simulations suggest that more than half of the FG-repeats of NPC finger proteins are actually exposed on the bundle surface and are not involved in contact with any other FG-repeats within 8 Å . Moreover, the simulation also suggest that the finger proteins adopt a brush-like structure. In this regard our results support the virtual-gate model. AFM experiments also suggest a similar brush-like behavior of a human FG-nup, namely Nup153
(
). The main difference between these models
is the degree to which FG-FG binding is involved in forming the gating barrier in the NPC. Our simulations suggest that more than half of the FG-repeats of NPC finger proteins are actually exposed on the bundle surface and are not involved in contact with any other FG-repeats within 8 Å . Moreover, the simulation also suggest that the finger proteins adopt a brush-like structure. In this regard our results support the virtual-gate model. AFM experiments also suggest a similar brush-like behavior of a human FG-nup, namely Nup153
(![]() ,
, ![]() ).
The many exposed FG-repeats seen on the bundle surface are also consistent with the reduction-of-dimensionality model
(
).
The many exposed FG-repeats seen on the bundle surface are also consistent with the reduction-of-dimensionality model
(![]() ),
which proposes that transport receptors pass through the NPC by sliding along the FG-surface. In fact, a sliding motion of NTF2 due to dynamic binding and unbinding involving two PHEs of one FG-repeat was observed in our 60 ns simulation.
),
which proposes that transport receptors pass through the NPC by sliding along the FG-surface. In fact, a sliding motion of NTF2 due to dynamic binding and unbinding involving two PHEs of one FG-repeat was observed in our 60 ns simulation.
Our results are summarized in the figure below. Several movies demonstrate the collapse of the stretched out FG-nups and show in further detail the structures of the bundle-based brush-like structure suggested by our simulations.
An array of individually tethered nsp1 segments was constructed and simulated
to sample computationally a representative volume of the nsp1-filled NPC central channel
by combining coarse-grained and all-atom molecular dynamics.
More Movies
 |
Movie 1. The formation of a wild-type brush-like structure (wild-type1 mpg 8.7M).The initially fully extended segments coiled and shortened quickly coming to a dynamic equilibrium. Each segment is depicted in a different color and the constrained N-termini (tethering points) are represented by red spheres. The formation of another wildtype brush-like structure and a mutant structure can also be found here: (wild-type2 mpg 9.2M), (mutant mpg 9.6M). |
 |
Movie 2. Detailed 360-degree view of the brush-like structure at the end of one wild-type array CG simulation (wild-type1 mpg 4.6M).Each segment is shown in a different color and the constrained N-termini are depicted as red spheres. The segments form bundles of 2 - 6 individual segments; different bundles exhibit inter-crossing of nsp1 segments. Detailed 360-degree view of another wildtype brush-like structure and a mutant structure can also be found here: (wild-type2 mpg 5.2M), (mutant mpg 5.4M). |
 |
Movie 3. Detailed 360-degree view of one final wild-type AA brush-like structure in surface representation (wild-type1 mpg 4.7M).The FG-repeats are colored with PHE in orange and GLY in green. Many FG-repeats are exposed on the surface of the structure, offering energetically favorable interaction spots to transport receptors. Detailed 360-degree view of another wildtype AA brush-like structure in surface representaion can also be found here: (wild-type2 mpg 5.3M). |
 |
Movie 4. The bindings to NTF2 (mpg 8.5M).Each nsp1 segment is colored in a different color and is shown in transparent. NTF2 is colored in red with its original position shown in gray. PHE343 and PHE341 of segment D16, and PHE360 of segment D15 are colored in orange. Closer views of the bindings are shown in Movie 4 (mpg 4.1M) and Movie 5 (mpg 3.3M). |
 |
Movie 4. The process of binding to NTF2 binding spot 2 for PHE343 and PHE341 (both of segment D16) (mpg 4.1M).PHE343 and PHE341 are shown in orange, and segment D16 in yellow. The exchange of binding between PHE343 and PHE341 resulted in a motion of NTF2 along the direction of segment D16; this motion can be clearly recognized in the movie. Ruler grids are also shown in the movie, with each grid being 10 Å X 10 Å . |
 |
Movie 5. The process of binding to NTF2 binding spot 1.3 for PHE360 of segment D15 (mpg 3.3M).PHE360 and PHE362 is shown in orange, and segment D15 in yellow. PHE360 was drawn towards NTF2 from further away during the simulation. Ruler grids are also shown in the movie, with each grid being 10 Å X 10 Å . |
Publications and News
-
Publications Database Transport-related structures and processes of the nuclear pore complex studied through molecular dynamics. Lingling Miao and Klaus Schulten. Structure, 17:449-459, 2009. -
Publications Database Cse1p binding dynamics reveal a novel binding pattern for FG-repeat nucleoporins on transport receptors. Timothy A. Isgro and Klaus Schulten. Structure, 15:977-991, 2007. -
Publications Database Association of nuclear pore FG-repeat domains to NTF2 import and export complexes. Timothy A. Isgro and Klaus Schulten. Journal of Molecular Biology, 366:330-345, 2007. -
Publications Database Binding dynamics of isolated nucleoporin repeat regions to importin-β. Timothy A. Isgro and Klaus Schulten. Structure, 13:1869-1879, 2005. See a review of our recent work in the journal Structure.
See another review of our earlier work in Structure, as well as a review in the journal Science in their feature Editors' Choice.
Our work was featured as a success story for computing at both the Pittsburgh Supercomputing Center and the National Center for Supercomputing Applications.
Our work was also featured at WhyFiles.org, a website dedicated to accessible explanations of scientific research for the novice scientist and layperson.
Investigators
- Lingling Miao
- Tim Isgro
- Yi Wang
- Klaus Schulten
- Murray Stewart (University of Cambridge)
- Andrej Sali (UCSF)
- Mike Rout (The Rockefeller University)
- Reiner Peters (Münster University)
Page created and maintained by Tim Isgro and Lingling Miao.



