Cells' Get Sticky with Calcium
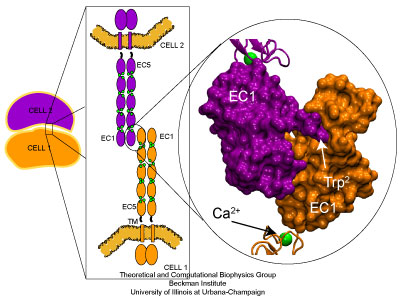 Cadherin molecules mediate calcium dependent cell-cell adhesion.
Cadherin molecules mediate calcium dependent cell-cell adhesion.
Adhesion between human cells organizes our body into its organs and parts. The adhesion comes about through an intricate system of molecules that perform their task in a highly selective manner such that the body's different types of cells will find the right cells and stick to them. This selectivity leads to tissue differentiation and the organization of organs as complicated as the brain. Cadherin proteins form a particular family of such adhesion molecules. Interestingly, they glue cells together only in the presence of calcium. Some members of the cadherin family of proteins are also involved in the transduction of sound and cadherin mutants are known to cause hereditary deafness (see the April 2005 highlight, "Hearing: Turning Sound into Voltage"). How cadherins selectively bind to each other and the role of calcium was not well understood, but now molecular dynamics simulations have offered magnificent insight into calcium's role. The simulations took advantage of parallel supercomputers and NAMD's ability to harness their power. The simulations revealed that in the absence of calcium cadherin behaves as a loose and weak rope. In the presence of calcium, however, cadherin molecules turn into stiff hooks that link cells together. The simulations also revealed that key residues, similar to those involved in hereditary deafness, interact with calcium ions to protect cadherin from unfolding when large mechanical forces are applied.
Influence of Ions on C-Cadherin Equilibrium Dynamics
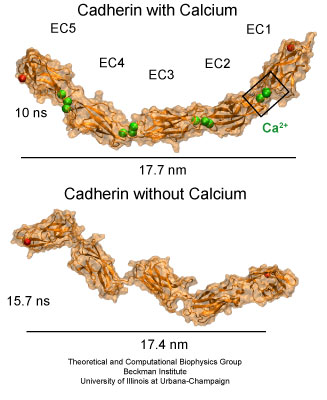
The extracellular domain of C-cadherin features five repeats with calcium ions binding at linker regions between repeats. The domain adopts a stiff, yet elastic ordered shape in presence of calcium ions. A disordered conformation is achieved upon removal of calcium.
Molecular dynamics simulations were carried out using the complete extracellular domain of C-cadherin, a Xenopus cadherin involved in the processes that drive morphogenesis during amphibian gastrulation. The simulated system included about 350,000 explicit atoms of water molecules and protein. The coordinates of the protein atoms were obtained from the x-ray crystal structure of C-cadherin. The protein features five cadherin repeats, each made of about 110 amino acids and folded in a "Greek key" motif characterized by seven beta-strands forming two beta-sheets. Four sets of simulations of this structure were performed. The first set utilized a system containing the protein, water molecules, and 12 crystallographically resolved calcium ions. The second set was performed without calcium ions. The last two sets of simulations involved systems in which crystallographically resolved calcium ions were replaced by potassium or sodium ions.
The equilibrium simulations of C-cadherin revealed that the extracellular domain of this protein maintains its shape and rigidity when calcium binding motifs at linker regions are fully loaded with this divalent ion. However, removal of calcium led to deformed conformations arising from the independent motion of individual repeats. Replacing calcium by potassium and sodium revealed a less drastic effect on the shape adopted by the protein, although the monovalent ions started to unbind from cadherin's calcium binding sites after a few nanoseconds of simulated dynamics. The outcome of the equilibrium simulations agree well with previous experimental results indicating that calcium rigidifies the extracellular cadherin domain. The simulations also clearly show inter-repeat motion that could lead to a collapsed conformation in the absence of calcium.
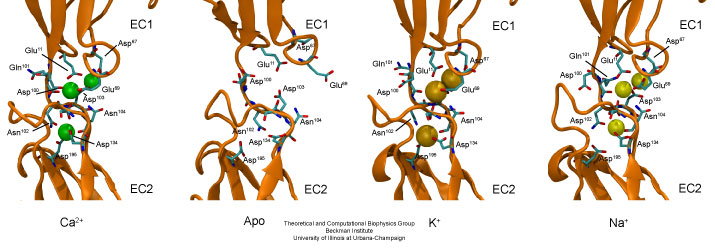
Linker regions between cadherin repeats feature calcium binding motifs. Simulations studied the behavior of cadherin in the presence and absence of calcium, as well as in the presence of potassium or sodium ions replacing calcium.
Tertiary and Secondary Structure Elasticity of C-Cadherin
The stability of cadherin was further probed by using conformations from equilibrium simulations as the starting points for constant-velocity steered molecular dynamics simulations. The SMD simulations were performed on systems with and without calcium, and on a system in which sodium replaced the crystallographically resolved calcium ions.
The SMD simulations revealed that upon stretching, the complete C-cadherin extracellular domain becomes straight by rearranging the relative orientation of individual repeats with respect to each other. This so-called tertiary structure elasticity was observed here in SMD simulations of C-cadherin with and without calcium, and with sodium replacing calcium. While the simulations performed in the absence of calcium show that linkers between repeats extend fairly easily, the simulations in which cadherin was loaded with calcium show that these ions act as molecular bearings, such that linkers regions behave as stiff hinges and the extracellular domain responds as one unit. The simulations in fact suggest that tertiary structure elasticity for cadherin with and without calcium has different molecular origins. The tertiary structure elasticity observed for cadherin without calcium seems to be rather entropic in origin, while elasticity of cadherin with calcium is reversible and non-entropic.
While unfolding of individual domains may not play a physiological role in adhesion, we explored the effect of calcium and sodium on the stability of individual repeats when subject to large forces inducing mechanical unfolding. The SMD simulations showed what is perhaps the most dramatic difference between the elastic response of the protein with or without calcium. While the EC1 repeat unfolded fairly easily in the absence of calcium, unfolding of EC1 in the presence of calcium required a considerably larger force that breaks a bond between a negatively charged amino-acid and a calcium ion.
Previous SMD simulations identified two types of mechanical unfolding processes for immunoglobulin-like domains in which water mediates rupture of hydrogen bonds concertedly (sheering mode) or one-by-one (zipper mode). In both types of unfolding, rupture of hydrogen bonds between beta-strands (concertedly or one-by-one) gives rise to the unfolding force-peak. Now there is a third mechanical unfolding category in which a bridge between charged amino-acids and divalent ions produce the unfolding force-peak. Thus, calcium is not only acting as a molecular bearing that facilitates cooperative motion of cadherin repeats, but also as a staple that maintains the stability of the structure providing further resistance to mechanical unfolding.
Simulations suggest then, that conserved residues involved in calcium binding are essential for mechanical stability of cadherin molecules; such residues when mutated may weaken cadherin molecules, thereby explaining why mutations at calcium binding sites of some cadherin molecules cause hereditary deafness.
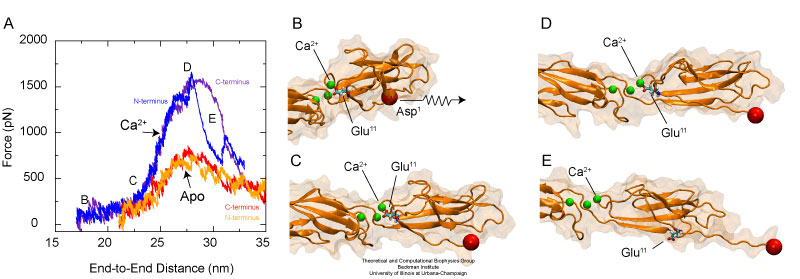
The elastic behavior of cadherin depends dramatically on the presence or absence of calcium ions. Unfolding occurs after a bridge between conserved charged amino-acids and calcium ions is disrupted.
- Click here for a movie (mI, mpeg, 1.6M) showing the dynamics of C-cadherin in the absence of calcium. The protein is shown throughout a 5 nanoseconds equilibration, followed by 0.65 nanoseconds of stretching, and subsequent relaxation lasting 5 nanoseconds. The EC1 repeat has been aligned to itself along the trajectory so as to illustrate the motion of repeats EC2-EC5 with respect to EC1. The trajectory reveals significant mobility of repeat EC1.
- Click here for a movie (mII, mpeg, 2.0M) showing the dynamics of C-cadherin in the presence of calcium ions. The protein is shown throughout a 5 nanoseconds equilibration, followed by 0.67 nanoseconds of stretching, and subsequent relaxation lasting 5 nanoseconds. The EC1 repeat has been aligned to itself along the trajectory so as to illustrate the motion of repeats EC2-EC5 with respect to EC1. The trajectory reveals limited motion of EC1.
- Click here for a movie (mIII, mpeg, 5.6M) showing local deformations of C-cadherin in the presence of calcium ions (same simulation as in mII). C-cadherin is shown in cartoon representation and colored according to average strain. The trajectory reveals that most of the protein strain is localized at repeats EC3, EC4, and EC5 (mainly at linker regions).
- Click here for a movie (mIV, mpeg, 5.8M) showing local deformations of C-cadherin in the absence of calcium ions (same simulation as in mI). The trajectory reveals that most of the protein strain in the absence of calcium is localized throughout all linker regions.
- Click here for a movie (mV, mpeg, 485K) showing unfolding of C-cadherin in the absence of calcium. The protein is first straightened through extension of linker regions and then further extension is achieved through unfolding of EC1.
- Click here for a movie (mVI, mpeg, 547K) showing unfolding of C-cadherin in the presence of calcium, which act as molecular bearings allowing straightening of the protein followed by unfolding of repeat EC1.
- Click here for a movie (mVII, mpeg, 1.4M) showing a detailed view of C-cadherin unfolding in the presence of calcium. Unfolding occurs after a bridge between a negatively charged residue and calcium is broken.
Calcium Allosteric Control of Residues Involved in Cell Adhesion
The dynamics of the extracellular domain, controlled by calcium, determines the availability of repeat EC1 to form adhesive contacts with proteins coming from neighboring cells. There are also subtle changes in C-cadherin dynamics controlled by calcium that will affect its adhesive properties. For instance, during the simulations in which calcium ions were present, the side chain of a tryptophan residue essential for adhesion remained partially exposed at all times. However, when calcium ions were removed or replaced by monovalent ions, the EC1 domain was more mobile and the sidechain of the tryptophan residue fluctuated between two states: exposed, and partially buried.
The dynamic switching of the tryptophan side-chain availability strongly support experimental work using antibodies and independent mutagenensis experiments combined with force measurements suggesting that the availability of this amino-acid is allosterically modulated by calcium.
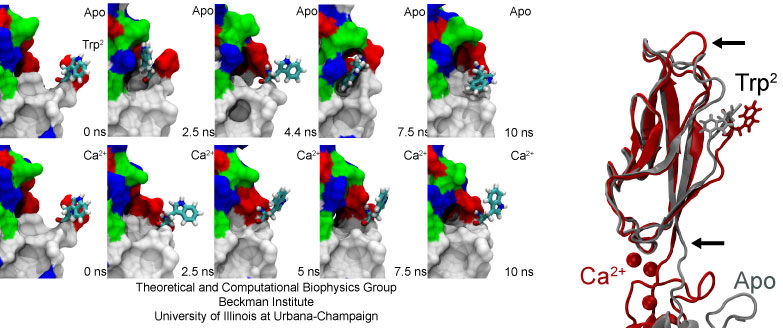
The availability of a tryptophan residue essential for adhesion is modulated by calcium ions. The side chain of Trp2 intermittently hides in the absence of calcium (Apo).
- Click here for a movie (mVIII, mpeg, 1.9M) showing the dynamics of a tryptophan residue essential for adhesion simulated in the absence of calcium.
- Click here for a movie (mIX, mpeg, 1.8M) showing the dynamics of a tryptophan residue essential for adhesion simulated in the presence of calcium.
- Click here for a movie (mX, mpeg, 1.9M) showing the dynamics of a tryptophan residue essential for adhesion simulated in the presence of potassium ions.
- Click here for a movie (mXI, mpeg, 1.8M) showing the dynamics of a tryptophan residue essential for adhesion simulated in the presence of sodium ions.
Publications
Tightrope act. David P. Corey and Marcos Sotomayor. Nature, 428, 901-902, 2004.
Investigators
Related TCB Group Projects
- Computational Force Microscopy
- Molecular Basis of Hearing
- Mechanosensitive Channel MscS
- Mechanosensitive Channel MscL
- Mechanical Proteins
-
See also related animation at 3Dciencia.
Page created and maintained by Marcos Sotomayor.



