TCBG Papers and Presentations at Supercomputing 2014
|
|
TCB Group Papers:
HPTCDL - First Workshop for High Performance Technical Computing in Dynamic Languages
*** Finalist, SC'14 Visualization and Data Analytics Showcase
Parallel Programming Laboratory Papers and Events
Members of the Parallel Programming Laboratory in the UIUC Department of
Computer Science are presenting six papers at SC'14.
NVIDIA Exhibition: VMD Interactive Ray Tracing
ray tracing, streaming from a remote GPU visualization
cluster located thousands of miles away in Santa Clara, CA.
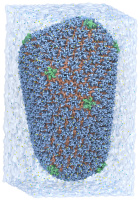
The demo will highlight all-atom HIV and chromatophore
structures simulated on the NSF Blue Waters supercomputer.
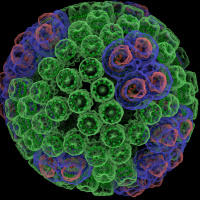
NVIDIA is demonstrating the world's first complete chemical structures of the HIV capsid and the chromatophore vesicle from purple photosynthetic bacteria, an organelle that converts light to energy.
This ray traced, interactive visualization allows researchers to analyze the HIV capsid and chromatophore down to the atomic level. The demo shows an interactive VMD molecular visualization powered by NVIDIA technologies, including the OptiX ray tracing library, and CUDA. The NVIDIA visualization servers are located thousands of miles away to show that scientists can interactively visualize their data remotely, eliminating the need to move large files from supercomputers to local workstations. Interactive ray tracing provides high quality lighting and transparency, that help researchers use their visual intuition to explore complex 3-D atomic structures and their spatial relationships.
The VMD software, and the HIV and chromatophore structures on display were produced by the Theoretical and Computational Biophysics Group at the University of Illinois at Urbana-Champaign. The chromatophore visualization is also nominated as one of the six finalists for the SC'14 Visualization and Data Analytics Showcase Award.



