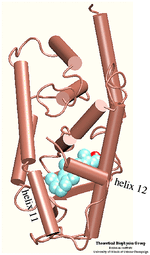
Ligand Binding Domain
Nuclear hormone receptors are ligand-activated transcription factors that regulate gene expression by interacting with specific DNA sequences upstream of their target genes. The mechanism of action for these receptors involves as a first step activation through binding of the hormone and as a second step the receptor binding to DNA.
The domain responsible for hormone binding is the ligand binding domain (LBD). This domain participates in several activities including hormone binding, homo- and/or heterodimerization, formation of the heat-shock protein complex and transcriptional activation and repression. The binding of the hormone induces conformational changes that seem to control these properties and influence gene expression. The conformational changes that accompany the transition between the liganded and unliganded forms of the nuclear hormone receptors affect dramatically their affinity for other proteins. In order to understand the functional role of the hormone one must understand the binding mechanism by which the hormone induces conformational changes.
Which residues are most involved in the process of binding/unbinding? How large are the conformational changes induced in the protein by the binding/unbinding process? Can the hormone use one pathway for binding and another pathway for unbinding? To answer these questions, the unbinding of retinoic acid from its receptor was induced by applying an external force on the hormone.
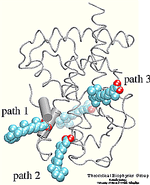
The crystal structure of the human retinoic acid receptor (hRAR)-gamma bound to all-trans retinoic acid (t-RA) suggests that the possible entry point for the hormone is closed by helix 12. Renaud et al. proposed that when t-RA binds to the receptor, helix H12 changes its conformation from extended into solvent, as seen in the crystal structure of retinoid X receptor (RXR), to a conformation closing the entrance to the binding pocket as in the crystal structure of retinoic acid receptor (RAR). Analysis of the surface of the protein identified another possible entry point: the only ``window'' in the surface of the protein that allows the hormone to be seen. Three possible unbinding pathways were studied. Path 1 and path 2 were chosen in the proximity of helix H11 and H12. Path 3 was chosen to study the unbinding of the hormone through the ``window''.
Simulation results indicated that it is possible to unbind the hormone along path 1 and path 3 without greatly affecting the structure of the protein. Particular characteristics of the simulated pathways suggest path 1 as the binding pathway for the hormone and path 3 as the unbinding pathway . Using the reversed order of events observed in the simulated unbinding along path 1 one can describe the binding mechanism of the hormone. First, Arg413, and then Arg396 attract and orient the carboxylate end of the hormone towards the binding pocket. When the hormone is within hydrogen bonding distance of these two residues, it experiences also the influence of the charged and polar residues Arg278, Lys236, Arg274, and Ser289 located at the opposite end of the binding pocket.
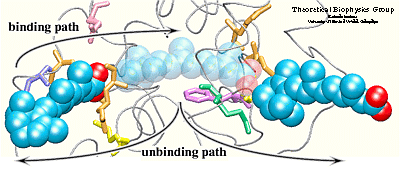
The two steps described above take place almost simultaneously, i.e., the strong electrostatic attraction between the carboxylate end of the hormone and residues Arg278, Lys236, Ser289, and Arg 274 helps the beta-ionone ring to pass between protein residues, thus, leading to the penetration of the hormone into the binding pocket. The entrance of path 1 is surrounded by highly fluctuating residues. These residues may become more ordered upon binding of the hormone, making contacts with the hormone or with other protein residues. Induced ordering of the protein side chains may be favorable for the hormone entry without requiring a motion of helix H12. Path 3 is an unlikely candidate for a binding pathway for the following reasons: 1) the carboxylate end of the hormone should be the one to be attracted by the point of entry into the protein since it furnishes stronger and more specific interactions than the beta-ionone ring; 2) the ``window'' would need to be sufficiently large to let the bulky ring pass through first. Therefore, one may then consider path 3 as a possible unbinding pathway, either as the sole unbinding pathway or an alternative to path 1.
Movies for the SMD simulations
- Unbinding along path 1 (1.6M)
- Unbinding along path 2 (1.6M)
- Unbinding along path 3 (1.6M)
Publications
Investigayors
References
- B. F. Luisi, W. X. Xu, L. P. Freedman, K. R. Yamamoto, and P. B. Sigler. Crystallographic Analysis of the Interaction for the Glucocorticoid Receptor with DNA. Nature 352:497-505, 1991
- M. G. Parker, editor. Nuclear Hormone Receptors. Academic Press, Sad Diego, CA, 1991.
- Ann M. Nardulli and David J. Shapiro. Binding of the estrogen receptor DNA-binding domain to the estrogen response element induces DNA bending. Molecular and Cellular Biology, 12:2037-2042, 1992
- M. Sabbah, S. Le Ricousse, G. Redeuith and E. Baulieu. Estrogen Receptor-Induced Bending of the Xenopus Vitellogenin A_2 Gene Hormone Response Element. Biochemical and Biophysical Research Communications, 185(3):944-952, 1992
- Helmut Heller, Helmut Grubmüller, and Klaus Schulten. Molecular Dynamics Simulation on a Parallel Computer. Molecular Simulation, 5:133-165, 1990
- Brad Banko and Helmut Heller. User Manual for EGO - Release 1.1. Theoretical Biophysics Group at the University of Illinois at Urbana-Champaign, Beckman Institute, 405 N. Mathews Ave., Urbana, IL~61801, U.S.A., 1991. [Beckman Institute Technical Report TB-92-07]
- J. W. R. Schwabe, L. C. Chapman, J. T. Finch, D. Rhodes and D. Neuhaus. DNA recognition by the oestrogen receptor: from solution to the crystal. Structure 15:187-204, 1993
- Board, Jr., John A. and J. W. Causey and Leathrum, Jr., James F. and Andreas Windemuth and Klaus Schulten. Accelerated Molecular Dynamics Simulation with the Parallel Fast Multipole Algorithm. Chem. Phys. Lett. 198:89-94, 1992