KcsA Potassium Channel
Ion channels are membrane-spanning proteins that form a pathway for the flux of inorganic ions across cell membranes. Diffusion of ions through the ion channels seems to be a simple physical process, and yet the ion channels are responsible for all electrical signaling in biology. Among their many functions, ion channels regulate the secretion of hormones into the bloodstream, generate the electrical impulses underlying information transfer in the nervous system, and control the pace of the heart and of the muscle system. To understand the physical basis of ion channel conduction, one needs to characterize the channel's structural and dynamic properties.
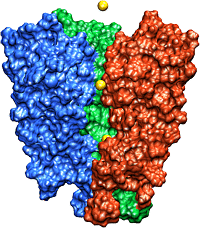
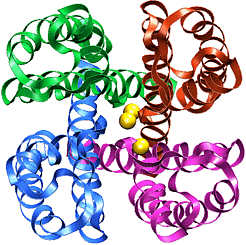
The recently solved structure of the K+ channel from Streptomyces lividans (KcsA channel) revealed that the KcsA channel is formed by four identical subunits, each subunit containing two alpha-helices connected by an approximately 30 amino acids long loop proofreading into the pore region. The pore region consists of an inner pore (starting from inside the cell), a large cavity near the middle of the pore, and the so-called selectivity filter that separates the cavity from the extracellular solution. The inner pore and the internal cavity are hydrophobic regions; in contrast, the selectivity filter is lined exclusively by polar main chain atoms belonging to the so-called "signature sequence". Experimental studies showed that this "signature sequence" is responsible for potassium selectivity. Advances made using structural and electrophysiological techniques have raised a new set of questions about the precise molecular details of ion selectivity. What is the chemical basis for the impressive fidelity with which the channel distinguishes between K+ and Na+ ions? How does the signature sequence encode a potassium selectivity filter? These questions are fundamentally structural in nature: therefore one has to "see" potassium ions coordinated at their binding sites in the pore before one can understand and explain ion selectivity in a potassium channel.
One of the best tools for making the connection between the structure and function of ion channels is molecular dynamics (MD) simulation, that allow one to follow conformational changes in the structure, and movement of K+ ions across the potassium channel. Unfortunately, full atomistic simulations of ion channels embedded in a lipid membrane have to account for thousands of atoms and the time scale of the permeation of even one single ion, of approximately 1 microsecond, is inaccessible to molecular dynamics simulations at present. The difficulty in simulating the passing of K+ or Na+ ions through the channel can be overcome by using Steered Molecular Dynamics (SMD) which "speeds" up a process. We propose to use SMD of the K+ channel to study the passing of K+ and Na+ ions through the channel. Trajectories obtained from the SMD simulations will allow us to determine the potential of mean force in both cases. We propose to test the hypothesis that the selectivity filter frees the K+ ion from its hydration shell more effectively than the Na+ ion. This should be reflected in the potential of mean force of K+ and Na+ ions along the axis of the channel, calculated by employing a time series analysis method. Ultimately, we seek to explain what is the basis for the selectivity of K+ ions over Na+ ions and how is this selectivity encoded in the signature sequence of the K+ channel.



