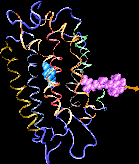
Retinal's Binding Pathway in bR
Formation of bacteriorhodopsin (bR) from the apoprotein and retinal has been studied experimentally, but the actual pathway, including the site of retinal entry, is little understood. Molecular dynamics simulations provide a surprisingly clear prediction. A window between bR helices E and F in the trans-membrane part of the protein can be identified as an entry site for retinal (purple van der Waals spheres in this figure). Steered molecular dynamics, performed by applying a series of external forces of 200--1000 pN over a period of 0.2 ns to retinal, allow one to extract retinal from bR once the Schiff base bond to Lys216 (blue van der Waals spheres) is cleaved. Extraction proceeds until the r etinal tail forms a hydrogen bond network with side groups lining the entry window. The manipulation induces a distortion with a fitted RMS deviation of coordinates (ignoring retinal, water, and hydrogen atoms) of less than 1.9 Angstroms by the time the entry window--retinal tail hydrogen bond network forms. The forces used are consistent with those expected for fast extraction and do not indicate significant potential barriers. We suggest that the path followed in the simulated extraction of retinal also describes the binding of retinal. The simulation reveals that movement of a water molecule between Lys216 and retinal's aldehyde group can initiate unbinding.
Publications
Binding pathway of retinal to bacterio-opsin: A prediction by molecular dynamics simulations. Barry Isralewitz, Sergei Izrailev, and Klaus Schulten. Biophysical Journal, 73:2972-2979, 1997.