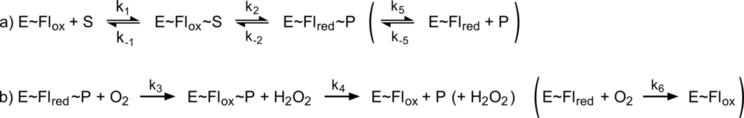
Oxygen migration in DAAO
Oxygen access channels - a general principle?
Molecular oxygen participates in numerous cellular processes but in most cases little is known about the mechanisms how oxygen reaches the reaction sites of the related proteins. However, in recent years specific oxygen access channels have been revealed for a number enzymes such as lipoxygenase, copper amine oxidase or glucose oxidase. In guiding oxygen to the catalytic site and by providing specific environmments for oxygen and the according reaction products these channels play significant roles in controlling O2 reactivity.
There are many more promising candidates for oxygen access channels, especially among monooxygenases and oxidases from the flavoprotein family. In these enzymes, where O2 (re)oxidizes the reduced flavin cofactor, key questions regarding the control of O2 reactivity with the reduced flavin cofactor are still open.
D-amino acid oxidase
Our work, a collaboration with Prof. Pollegioni's group at the University of Insubria, Italy, addresses these issues using D-amino acid oxidase (DAAO) [1] . The enzyme (EC 1.4.3.3) is found in most mammalian cells and catalyzes the oxidative deamination of D-amino acids to yield alpha-keto acids, ammonia and hydrogen peroxide. Even though it was already studied in the 1930s by Krebs and Warburg its physiological role is not yet fully understood. However, it has been connected with the degradation of the neurotransmitter D-serine in the brain and with the regulation of neurotransmission. Being a prominent member of the flavoprotein oxidase family, DAAO is regarded a key enzyme for understanding the flavin catalysis mechanism.
|
More details about the rection mechanism can be found here.
In DAAO the catalytic center is located at the end of a funnel serving as entrance for the substrate D-amino acid. In this channel the cofactor flavin is shielded by the bound D-amino acid substrate or by the product imino acid from access of small molecules originating from the solvent region.
Let's suppose there is a channel for O2 connecting solvent and the flavin pocket. But what makes a good O2 access channel? Ideally, it is a continuous region between solvent and the active site where the average probability of finding O2 is high. This probability depends on many factors like the dynamics of protein side chains and water molecules or the hydrophobicity of the environment.
Unfortunately, localizing dioxygen in proteins is difficult. Oxygen is very mobile and usually is not resolved in crystal structures and other techniques, like tryptophane fluorescence quenching, suffer from insufficient resolution. However, a promising approach to trace oxygen consists of implicit ligand sampling, a technique that allows efficient computation of 3D free-energy maps for small gas molecules in proteins.
ILS calculations reveal oxygen access pathway
|
Implicit ligand sampling (ILS) is an efficient method, based on molecular dynamics (MD) simulations, to compute the probability density for small, weakly interacting particles such as O2 at all positions in a molecular system [2]. From this probability useful quantities can be calculated: (i) Occupancy, the probability to find a particle in a certain volume region. (ii) Gibbs free energy costs ΔG(O2) of relocating O2 from a reference state (e.g. solvent) into a certain region. (iii) Affinity for O2 for a given region expressed as an apparent dissociation constant Kd,app.
Our approach combines the strengths of computer simulations and biochemical experiments. Using ILS we predicted high affinity regions for O2 inside the protein and in the vicinity of the flavin and extract the most likely diffusion pathways.
The ILS calculations are based on MD trajectories of the DAAO-imino pyruvate complex. They render the 3D distribution of the free energy potential ΔG(O2) of O2 relative to its potential in the bulk solvent. The lower the free energy for a given point in space, the higher is the affinity for O2 and therefore the average O2 occupancy.
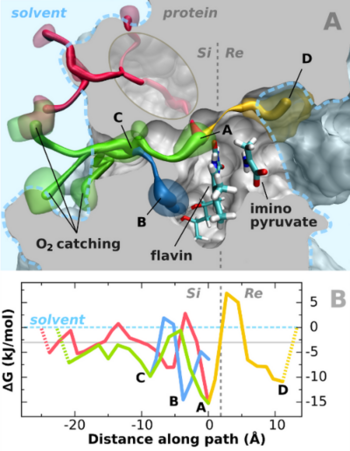
Fig. 2A shows ΔG(O2) in the vicinity of the active site in form of an isoenergy surface. On the Si-side (left) of flavin there are two high affinity regions inproximity of the cofactor: site A is adjacent to atoms N(5) and C(4a) while site B is close to the xylene edge of the flavin. Further high affinity sites are located just below the protein surface (site C) and in a funnel at the surface. Could these sites be parts of an access channel network for O2? In order to address this question we extracted the minimum energy paths connecting the O2 high affinity sites near flavin with the solvent. The results show that indeed the sites are connected via energetically preferable pathways which are displayed in Fig. 2A together with their respective energy profiles in Fig. 2B.
The energy level along the access channel towards site A, including the
barriers, is below the solvent level, while the path towards site B
exhibits maxima that are insignificantly higher than solvent level.
I.e. inside the channel, particularly at sites A and B in the vicinity of
flavin, the virtual O2-concentration is substantially higher than
that in an equivalent solvent volume thus speeding up the flavin reoxidation
process.
Note that the movement along the pathways
should be understood as guided, reversible
diffusion inside the channel system: consequently,
all sites along the pathways may be considered
connected. Since due to the low barriers and small
distances the diffusion time between any two
places inside the channel system can be assumed
much shorter than the mean time for the access of
another O2, the entire system contributes to the
increase of the O2 "effective concentration" at the
reaction center.
Obstructing O2 access - the Gly52X DAAO variants.
|
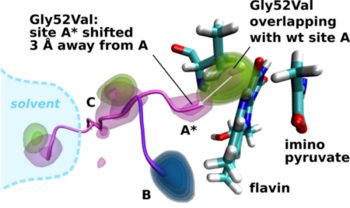
The O2 access hypothesis was tested computationally as well as experimentally. In case the the putative access channel does play a significant role in the enzyme reaction cycle then an attempt to block the O2 path should result in a reduced reactivity. Inspecting the 3D structure one notices that the additional side chain bulk of any mutation of residue Gly52 would interfere with high affinity site A (Fig. 3) by partially filling the space expected to be occupied by O2 during the electron transfer step. Consequently we prepared a number of Gly52X DAAO variants by site-saturation mutagenesis and investigated their biochemical properties with respect to O2 reactivity. The main experimental result is that the reactivity of the DAAO variants in the oxidative half-reaction is drastically lowered (up to 100 fold in Gly52Val-DAAO). The computational assessment of the Gly52Val substitution shows that in the variant the high affinity site A* has substantially less affinity for O2 and the energy minimum is shifted 3 Å away from flavin and the original site A (wt DAAO). At the same time site B remains unaffected by the mutation.
Implications for the reaction mechanism
Sites A and B both lie directly adjacent to the flavin. However, since the highest density of the reduced flavin negative charge is at positions C(4a)-N(5), site A would constitute an ideal location for efficient electron transfer to O2. The additional sites of O2 affinity (Fig. 1A) could also play relevant roles. Site C is located just under the protein surface at the entrance of the channel leading to the high affinity sites A and B. Together with the affinity sites at the protein surface it constitutes a region where O2 is "collected/concentrated" for further transport and utilization (catchment area). Once O2 has entered the channel system, it can move about freely (along the paths) visiting the reaction site frequently. The O2 equilibration between channel and solvent is much faster than the reaction time. Hence, many O2 molecules enter and leave the channel before eventually one of them participates in the reaction. The affinity of the channel system increases the probability for each O2 to stay close to flavin thus enhancing its bimolecular reaction term.
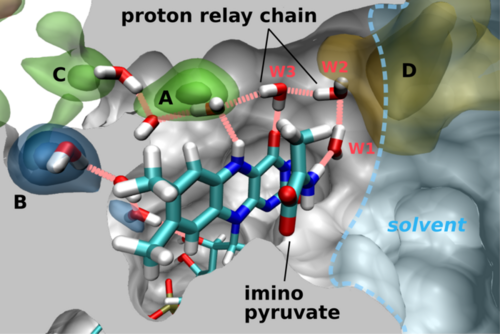
Protein dynamics reveals a proton relay system
|
Comparing the X-ray structure with the MD trajectories yields
interesting clues. No H2O is observed at site A in
any of the four different DAAO X-ray structures.
In the simulations, however, a transient presence of water molecules
can be observed. In fact the putative O2 channel
is flexible and the H2O molecules inside are not fixed but in
constant exchange with the bulk water phase. Throughout the MD
trajectories H2O molecules diffuse in and out of all high
affinity sites. A lone H2O in the hydrophobic cavity A is not
favorable but a H-bond chain connecting it to hydrophilic regions can
significantly stabilize H2O in that position. Notably, during
parts of the simulation there exists a contiguous chain of 4 H-bonded
H2O molecules crossing the flavin plane between the imino
pyruvate and site A that might be used as H+ relay system.
Its role could be the transfer of a H+ from the =NH2+ function of bound imino pyruvate to the developing, negatively charged O2 species. This H+ relay is shown in Fig. 4 and 5.
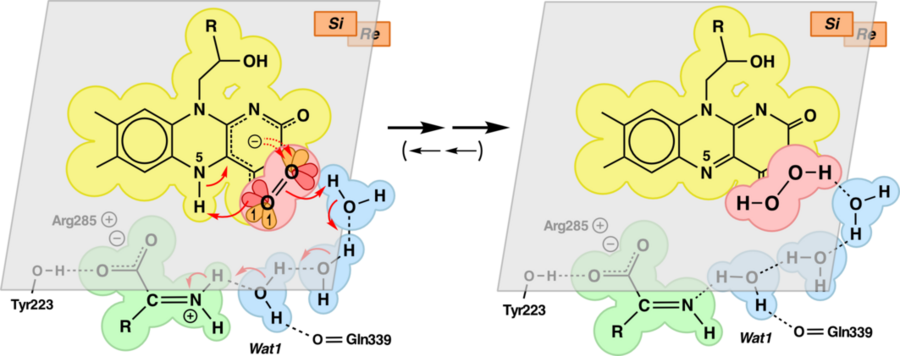
|
Publications
[1] O2-reactivity of flavoproteins: Dynamic access of dioxygen to the active site and role of a H+ relay system in D-amino acid oxidase. Jan Saam, Elena Rosini, Gianluca Molla, Klaus Schulten, Loredano Pollegioni, and Sandro Ghisla. Journal of Biological Chemistry, 2010. In press.
[2] Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Jordi Cohen, Anton Arkhipov, Rosemary Braun, Klaus Schulten Biophysical Journal, 91:1844-1857, 2006
Investigators
UniversitÓ dell┤Insubria, Italy:- Sandro Ghisla
- Gianluca Molla
- Loredano Pollegioni
- Elena Rosini



