Gas Transport inside hydrogenase CpI
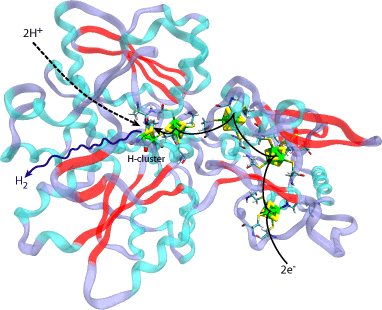
Fig 1. The structure of CpI Hydrogenase from Clostridium pasteurianum with its naturally embedded metallo-clusters. Shown with arrows are the pathways for the electrons, hydrogen ions, and the hydrogen product to and from the active H-cluster.
The promise of cheap renewable energy
Hydrogenases constitute a family of enzymes found in certain microorganisms, which catalyzes the reversible oxidation of hydrogen gas. While the host organisms of a hydrogen-producing hydrogenase, such as CpI hydrogenase in Clostridium pasteurianum, will typically use it to dispose of excess electrons, many scientists are interested in the enzyme for a completely different reason. In fact, cultures of hydrogenase-containing microorganisms have the ability to produce a constant output of hydrogen gas (H2) from just sunlight and water. Electrons from a carrier combine with hydrogen ions from the cell environment at the hydrogenase's buried H-cluster active site to produce H2 (Fig 1). If harnessed properly, hydrogenase and/or hydrogenase-containing organisms could be used to supply affordable and renewable H2 to be used as an energy fuel, and thus solve the "supply" aspect of the future hydrogen economy.
This idealistic picture is not without problems. Notably, hydrogenase's H-cluster is extremely sensitive to the presence of oxygen gas (O2), which will bind to it permanently. In the presence of O2, hydrogen production is maintained for only a few minutes before the hydrogenases become deactivated. In order to maintain a sustained hydrogen production using hydrogenase, an anaerobic environment is currently required, making hydrogenase a costly and impractical source of H2. This creates an interesting scientific problem: if we can identify the pathways through which O2 reaches the H-cluster, we could then create an engineered version of hydrogenase in which these O2 pathways are blocked, thus decreasing hydrogenase's sensitivity towards O2.
Swallowed by a protein: the fantastic voyages of O2 and H2
In order to understand how O2 is transported across hydrogenase to reaches the H-cluster, we generated an all-atom simulation model based on the PDB crystal structure of CpI hydrogenase (1FEH). We then investigated the diffusion of O2 and H2 inside the protein by placing copies of either H2 or O2 right at the H-cluster where the gas molecules bind. This system was then let go, and the motion of the gas molecules was monitored. To speed up the calculation, a computational trick called locally enhanced sampling (LES) was used. In our implementation of LES, instead of one copy of each gas molecule, we placed 1000 simultaneous copies which each interact with the hydrogenase protein with an interaction scaled by 1/1000, but which do not see each other. This enables us, in effect, to simulate 1,000 gas diffusion event simultaneously and get a much better picture of the gas molecules' movements inside the protein, than we would if we only simulated one gas molecule.
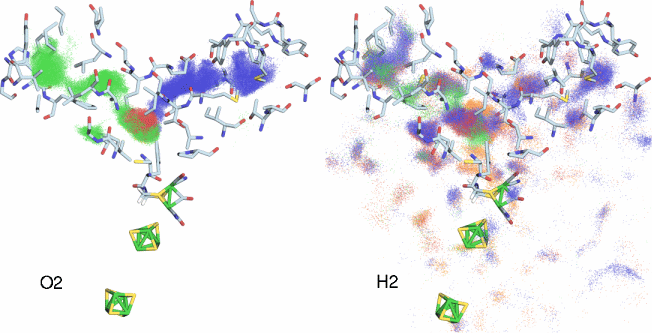
Fig 2. Simulation of O2 and H2 diffusion inside CpI hydrogenase. Each dot represents the position of a gas atom during the course of the simulations. The various dot colors (blue, green, red, orange) represent different independent diffusion simulations. Only the protein residues that line the major gas pathways have been drawn as licorice. This figure highlights the very different modes of diffusion for H2 and O2 inside hydrogenase.
What we discovered was surprising. First of all, both O2 and H2 exited predominantly through not one pathway, as had long been suspected, but two! Also interesting is the fact that one of the pathways was not detectable in the static structure, while the other pathway could only be detected using a very small probe radius. Furthermore, while our 1,000 copies of H2 spread out through the protein after leaving the H-cluster, the O2 molecules would stay clumped together in a small cloud during their entire trajectories, though the actual pathway taken varied from one trajectory to the next (Fig 2). In fact, it can be concluded that there are no O2 permanent "channels" inside CpI hydrogenase, but rather that O2 merely jumps between a series of dynamically fluctuating pockets inside the protein (see our paper). H2, on the other hand, behaved very differently. Due to its smaller size, the protein appeared much more porous to H2 than O2, and H2 was observed to almost freely diffuse through both of the O2 pathways, and occasionally exited the protein using entirely new pathways.
No permanent channels
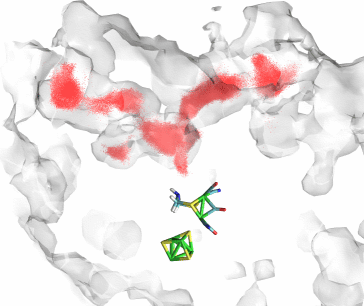
Fig 3. Predicted pathways for O2 based on the open space inside an equilibrium simulation of CpI in the absence of O2 (ghostly shell), along with the actual trajectories of O2 (red dots).
Various types of proteins often need to selectively recruit ligands into their interiors or across a membrane. Whether these ligands are ions, sugars, lipids or even water molecules, the proteins often exhibit cylindrical channels inside them to allow for the ligand's transport. For the case of CpI hydrogenase (and other gas-transporting proteins such as myoglobins, etc.), clear cut channel are hard to detect, even though gas transport across these proteins is clearly efficient. In the previous section, we alluded to the fact that O2 travels through a series of fluctuating cavities, or packing defects, present in the protein. If that is the case, this begs a few questions. Are all parts of the protein accessible to gas, or are certain areas predisposed to act as fluctuating channels? Do gas molecules interact with the protein in order to open it, or are the gas pathways somehow pre-defined inside the protein matrix? In our gas diffusion study, we detected two major O2 pathways, but could there be more pathways that we haven't detected?
To answer these questions, we developed a methodology for predicting gas pathways inside a protein by analyzing the dynamically fluctuating cavities that naturally arise inside a protein. By looking at an entire equilibrium trajectory of CpI in the absence of gas, we mapped, on a grid, all the areas of the protein which could naturally fit a particle the size of O2 or H2 at any point during the entire trajectory. Thus, we hypothesized that O2 (or H2) could only diffuse inside the protein where space is already made available for it. It turns out that this method predicted the location of the two gas channels as well as of other cavities where we observed high concentrations of H2, with very good accuracy (Fig 3). In conclusion, for the case of CpI, despite the lack of permanently open gas channels, the pathways taken by gas molecules such as O2 and H2 are predefined to lie in areas of the protein which have a natural disposition towards greater density fluctuations.
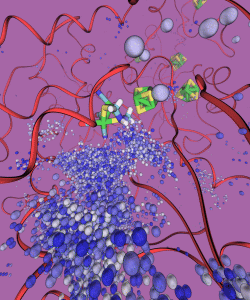 Depiction of the trajectory of H2 diffusing out from the H-cluster.
Depiction of the trajectory of H2 diffusing out from the H-cluster.
Conclusion
Our results demonstrate that gas pathways are often well-defined and can be predicted inside proteins despite the lack of obvious features such as channels. Also, we show that O2 transport is highly dependent on the internal variability of the protein's density, whereas H2 transport acts as if the protein were somewhat porous. These results will allow researchers to target O2 pathways inside CpI hydrogenase to increase its tolerance to the presence of O2 without disrupting the outward transport of H2.
Publications
Investigators
- Jordi Cohen
- Paul King
- Kwiseon Kim
- Michael Seibert
- Maria Ghirardi
- Klaus Schulten
Page created and maintained by Jordi Cohen.



