Energy Conversion in F0-ATPase
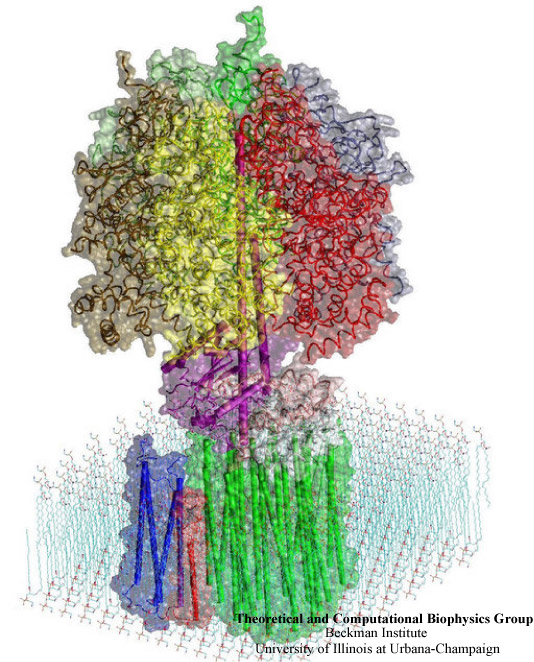
Adenosine triphosphate (ATP) is the primary energy "currency" in most living organisms. Storing readily-available energy in the form of a chemical bond, ATP delivers it to a reaction site and releases it in the process of hydrolysis, providing an energy supply for enzymatic reactions and biomechanical processes. ATP synthase is a large (about 100,000 atoms) protein, which includes a transmembrane F0 unit coupled to a solvent-exposed F1 unit via a central stalk gamma. The F0 unit utilizes a transmembrane electrochemical potential (proton motive force), converting it into the mechanical energy of the stalk rotation. The rotation leads to cyclic conformational changes in the catalytic sites in the F1 unit, thereby driving ATP synthesis. ATP synthase can also function in the reverse direction, hydrolyzing ATP and utilizing the released energy to pump protons across the membrane. This reversibility, along with the nearly 100% efficiency and the recently discovered, remarkably symmetric structure (1997 Nobel Prize in Chemistry), makes ATP synthase a perfect system for exploring interconversion between mechanical and electrochemical energy in molecular motors.
We seek to identify and explore the chain of the elementary chemical (proton transfer) and mechanical (domain motion) events involved in the process of converting the electrochemical energy of the transmembrane proton gradient into the mechanical energy of the c subunit oligomer rotation. The ultimate challenge is to understand how proton translocation across the membrane, which includes a sequence of local (~10A) proton transfer events, drives the rotation of the large (~70A) c subunit oligomer coupled to the central stalk.



