DBP5: Membrane Transporters
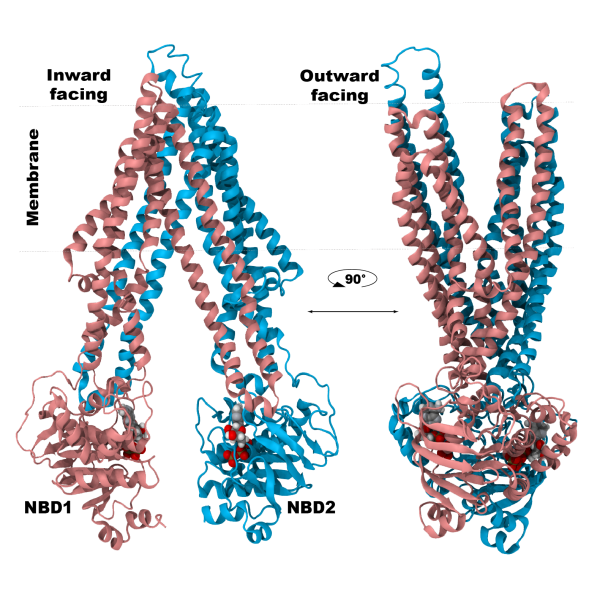
Membrane transporters perform the fundamental biological task of efficient and selective transport of materials across the cell membrane using diverse forms of cellular energy. Many transporters have been identified as drug targets, and effective drug development necessitates a comprehensive understanding of their structural intermediates and transitions during the transport cycle. The Center has developed a set of advanced methodologies combining extensive sampling of the phase space and free energy calculations to structurally and thermodynamically characterize the complete transport cycles of the transporters. Here, further development of this methodology will enable structural characterization of transport cycle of two structurally and mechanistically diverse classes of membrane transporters: (S1) P-glycoprotein (P-gp), a primary (ATP-driven) transporter implicated in multidrug resistance in cancer, and (S2) VcINDY, a secondary (ion-driven) transporter mediating transport of metabolic intermediates.
Describing large-scale structural transitions of membrane transporters in their native membrane environment in atomic detail is an extremely challenging problem. % for both experimental and computational structural biological studies. A primary challenge in non-equilibrium simulations, %required for describing slow and complex structural transitions underlying transport function, is the identification of optimal biasing protocols to drive the process. The Automated Reaction Coordinate Search (TRD1) tool will be essential to facilitate comprehensive combinatorial examination of system-specific collective variables that will be used to induce the transition and to dynamically evaluate the associated non-equilibrium work value, serving as a metric to assess the quality of the transition pathway. Once a transition pathway has been identified, calculation of the free energy profile along the pathway using enhanced sampling (TRD1) techniques will require 100s-1000s of copies of the membrane-embedded transporter system simulated/analyzed in parallel, which necessitates the efficient utilization of supercomputers by improving single-node enhanced performance (TRD1) as well as use of an improved version of TimeLine and parallel analysis (TRD2). A further challenge stems from the intricate protein-lipid interactions and slow diffusion of lipids, particularly ones intercalated between protein domains/helices that undergo very large reorientation during the transport cycle. This challenge will be addressed by the Mem(Re)Shaper (TRD3) tool, allowing one to use time-dependent, specifically crafted grid potentials to smoothly guide the lipids out of a defined space. Despite employing non-equilibrium simulations, given the mechanistic complexity of membrane transporters and their interactions with diverse lipids in the membrane, we will continue to heavily rely on long time-scale MD simulations, which will be accelerated by the single-node enhanced performance (TRD1) improvements. An additional challenge in (S1) is to describe and take into account the process of ATP hydrolysis. This challenge will be addressed by the QM/MM (TRD1) interface in NAMD.
The investigations in (S1) and (S2) will build upon the long tradition of the Center in studying mechanistically and structurally diverse membrane transporters, in particular upon the recent development of a novel methodolgy for transition pathway calculations, which has been successfully employed in describing large structural changes of ABC transporters, and in describing the full thermodynamic transport cycle of a secondary transporter. The first step will be to generate structural models of the end states (inward- and outward-facing states), which are necessary for the characterization of the transition pathway. Existing crystal structures of both Pgp and VcINDY provide structural information for only one state. The unknown states will be constructed using ModelMaker (TRD3) while integrating experimental data obtained from our collaborating labs (Mchaourab, Wang). The Automated Reaction Coordinate Search (TRD1) tool and enhanced sampling methods (TRD1) will be used to identify optimal biasing protocols, which in turn will be used to characterize the transition pathways in (S1) and (S2). These pathways will then be thermodynamically characterized using bias exchange umbrella sampling calculations yielding a free energy profile for the transition. The Mem(Re)Shaper (TRD3) tool will be employed to specifically guide intercalated lipids during the structural transition of the transporter in (S1) and (S2). EPR distance measurements (Mchaourab) describing dynamic characteristics of the protein will be used in modeling and transition path sampling. Mechanistic understanding of energy utilization in (S1) and (S2) will be building on the functional and kinetic data provided by our collaborators (Nakamoto, Wang).
