DBP4: Neuron and Synapse
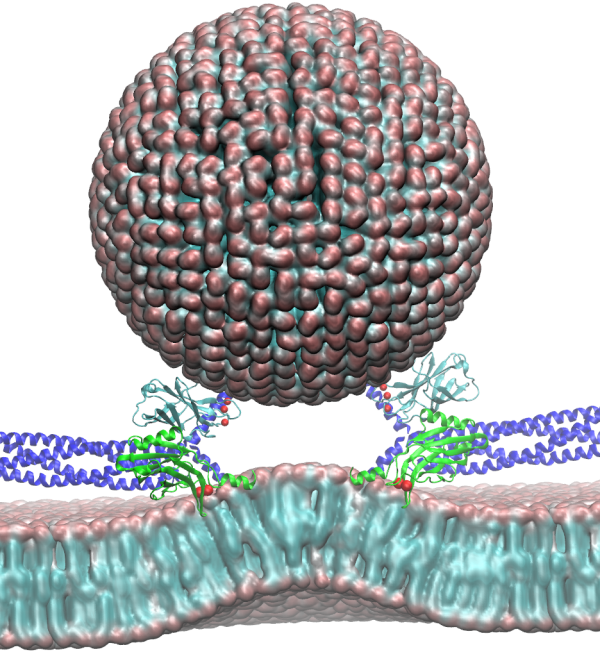
'Insert Caption Here'
The brain is the source of thoughts, perceptions, emotions, memories and actions. Neural signaling, the foundation of brain activity, must be precisely regulated to prevent neuronal disorders that may cause Parkinson's disease, schizophrenia, compulsive behaviors and addiction. Such a precise regulation is achieved by key signaling proteins, voltage-gated sodium (Na+) and potassium (K+) channels for electrical signaling and calcium (Ca2+) - bound synaptotagmin for chemical signaling. Here, innovations in computer simulation techniques will be used to investigate the molecular mechanism of (S1) neural firing induced by voltage-gated Na+ and K+ channels and (S2) membrane fusion triggered by synaptotagmin.
In (S1), a key challenge is the lack of a eukaryotic Na+ channel structure, which inhibits further understanding of its molecular mechanism on signal regulation. ModelMaker (TRD3), a hybrid modeling tool combining de novostructure prediction, molecular dynamics flexible fitting (MDFF) and homology modeling, will be developed to construct the atomistic channel structure, and its conformations at different conducting states. Transition rates between the channel's different conducting states are crucial to neural firing regulation, but the microsecond timescale channel action in response to membrane voltage changes becomes a bottleneck to estimate such transition rates. Single node enhanced performance (TRD1) in NAMD, aiming at simulation speed of one microsecond a day, will enable long simulations of the channel, a system of ~100 K atoms, and direct measurement of the channel's structural transition rate. In (S2), a fundamental challenge is that the membrane fusion process spans a wide-range of temporal and spatial scales, involving small-scale atomistic Ca2+ ion interactions, a large-scale membrane morphology change in fusion with a system size of ~100~M atoms, and a millisecond long time-scale fusion event. A multi-scale replica exchange (TRD1) algorithm in NAMD will be developed to simulate the system with atomistic resolution, while using a large number of coarse-grained PACE model replicas to enhance sampling efficiency and guide the atomistic simulation. In addition, a further challenge is that lipid motion is too slow to simulate a full fusion event, even in a coarse-grained representation. A Mem(Re)Shaper (TRD3) algorithm will be developed to apply time-dependent grid forces to the system to drive lipid reorganization towards fusion. Furthermore, in the atomistic replicas, divalent ions like Ca2+ are not well-described by conventional force fields, and atomic polarizability is needed for improvement. In response,atomic polarizability will be implemented in NAMD through the Drude polarizable force field (TRD1) for systems of more than 10M atoms taking advantage of performance enhancements in NAMD.
Our investigation of (S1) and (S2) will build upon previous work performed by the Center revealing mechanistic details in neurons and synapse inaccessible to experiments, including neural networks , voltage-gated K+ channels , synaptic proteins, and protein triggered membrane morphology changes. In (S1), homology modeling based on the resolved bacterial channel structures will be performed with ModelMaker (TRD3) to construct the eukaryotic Na+ channel conformations at different conducting states. Then structural transitions between the channel's conducting states will be resolved by microsecond-long simulations, taking advantage of improved NAMD performance (TRD1), guided by cross-linking experiments (Ahern). The resulting structural transition rates in response to membrane voltage, together with previously determined K+ channel structures and transition rates, will be incorporated into a molecular version of the Hodgkin-Huxley equation recently derived to model neural firing. In (S2), incorporating synaptotagmin crystal structures (Brunger) and FRET measurements (Shin), multi-scale replica exchange(TRD1) simulations with biasing forces from Mem(Re)Shaper (TRD3) algorithm will be performed to elucidate the dynamic process of membrane fusion triggered by Ca2+-bound synaptotagmin. Snapshots of important intermediate fusion stages will be selected and serve as initial coordinates for millisecond-long atomistic simulations employing the Drude polarizable force field (TRD1) and performance enhancements for large systems in NAMD. Synaptotagmin's role in regulation will then be characterized at different stages of membrane fusion, e.g., membrane bending, hemifusion, and fusion pore formation.
