DBP3: Molecular Motors
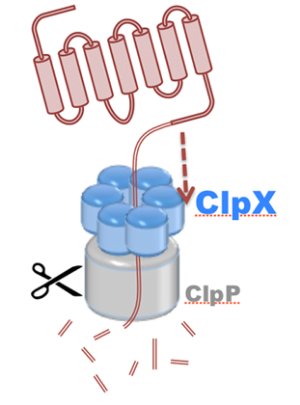
Motor proteins are molecular machines that convert chemical energy from ATP hydrolysis into mechanical work to drive conformational transitions regulating a range of biological processes. Defects in motor protein function are implicated broadly in cancer, as well as numerous cardiovascular, neurological, and reproductive diseases. The chemo-mechanical properties of motor proteins remain poorly characterized for the majority of systems, primarily due to the structural complexity of molecular machines, as well as the long (millisecond) timescales of their action. Here, innovations in computer simulation techniques will enable investigation of the mechanisms of hydrolysis-driven motion underlying (1) regulation of ATP synthesis by ATP synthase (S1), (2) protein degradation by ClpX unfoldase (S2), and (3) cytoskeletal cargo transport by cytoplasmic dynein (S3).
A key challenge in the computational investigation of motor protein mechanisms is presented by the long timescales of motor action. To meet this challenge, one needs to perform MD simulations of many replicas of systems with <1M atoms to identify the slow conformational transitions. Improvements in single-node enhanced performance (TRD1), as well as the implementation of enhanced sampling (TRD1) algorithms designed for sampling transition paths, e.g., dynamic importance sampling, will permit such simulations. Another challenge arises from the difficulty of identifying a reaction coordinate within structurally complex motor systems that can measure transition progress and characterize transition dynamics. A protocol for automated reaction coordinate search (TRD1) will facilitate the systematic reduction of the motor configuration space to a collective variables space through an approach based on steered MD simulations and non-equilibrium work measurements. A further challenge stems from the inability of purely classical MD simulations to couple the hydrolysis reaction in the motor catalytic site with the conformational transition of motor action explored by path sampling techniques. The necessity to simultaneously simulate chemical reactions and conformational transitions during path sampling will drive the development of a fast QM/MM (TRD1) interface to NAMD. Once a transition path has been identified, analysis of the path for kinetic and energetic properties presents a challenge. Internal support of the milestoning enhanced sampling (TRD1) method in NAMD will provide an effective, efficient strategy for calculating rate constants and free energies along transition paths. Finally, simultaneous analysis of the ensembles of trajectories produced by sampling transition paths also presents a challenge, which will be addressed through further development of the TimeLine and parallel analysis (TRD2) tool.
The investigation of motor protein mechanisms in (1), (2), and (3) will build upon the Center's previous work in developing and applying path sampling techniques to reveal the translocation of RNA by Rho helicase, the rotation of the V-type ATPase central stalk, and the alternating-access mechanism of phosphate ion transport by the glycerol-3-phosphate:phosphate transporter, as well as our experience in performing QM/MM studies of the catalytic domains. of F-type ATPase and PcrA helicase. The Center will explain, in atomic detail, the mechanisms underlying rotatory catalysis of an entire S1, given a model derived from high-resolution EM data (Rubinstein), polypeptide translocation through the central pore of S2, connecting and confirming stepsizes and rates obtained from single-molecule studies (Martin), and the power stroke cycle of the motor domain of S3, including allosteric communication and dynamical motions observed with fluorescence experiments (Goldman). The automated reaction coordinate search (TRD1) protocol will be used to simplify selection of collective variables to describe motor transitions, while enhanced sampling (TRD1) techniques, i.e. dynamic importance sampling, will be used primarily to predict initial transition paths. A QM/MM (TRD1) interface to NAMD will allow for direct coupling of the hydrolysis reaction with the conformational transition of motor action in S1, S2, and S3. The milestoning method will be employed to determine the rate constants and free enery profiles characterizing the transitions of S1, S2, and S3, and TimeLine and parallel analysis (TRD2) will facilitate simultaneous analysis of the trajectories produced by sampling their transition paths.
