DBP2: Symbiont Bacteria within the Human Body
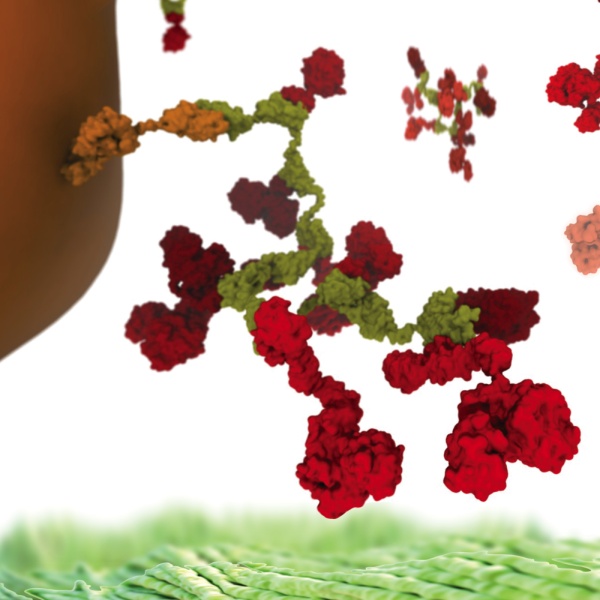
Symbiont bacteria greatly influence human health and play a significant role in pathogenesis, disease predisposition, physical fitness, and dietary responsiveness. Here we propose to investigate two key processes underlying bacterial interactions with humans: chemotaxis and plant fiber metabolism. Specifically, we will study the structure and function of the highly cooperative macromolecular complexes central to each of these processes, namely the chemosensory array (S1), a transmembrane cluster of sensory proteins, and the cellulosome (S2), a large extracelular enzymatic complex. Innovations in computer simulation techniques will be used to investigate, in particular, the molecular origins of (1) robust sensory signal transduction within (S1) and (2) highly efficient plant fiber degradation by (S2).
A central challenge for the investigation of (1) and (2) concerns the limited structural information available for individual proteins composing (S1) and (S2). In addition, due to the structural heterogeneity of (S1) and (S2), current experimental techniques are unable to resolve structures of the intact complexes at high resolution. High-throughput modeling techniques (TRD2), including a VMD interface to existing homology and de nov modeling tools such as Modeller and Rosetta, will be developed to resolve atomistic structures of (S2). In addition, ModelMaker (TRD3) will be used to obtain initial homology models for (S1) which will be refined using intermediate- and high-resolution electron microscopy (EM) data, driving MDFF advancements, including autoMDFF (TRD3) and the development of advanced density map segmentation and manipulation (TRD2) tools. An additional challenge for both (S1) and (S2) is the investigation of the functional dynamics occuring within these complexes over wide-ranging temporal and spatial scales. A fast semi-empirical QM/MM (TRD1) method taking advantage of powerful GPUs will permit the study of molecular events within (S1) and (S2) at both the single-molecule and electronic levels in the context of the larger complexes. Furthermore, new implementations of enhanced sampling (TRD1) methods, such as GSA in NAMD, will extend sampling of the intact complexes to the micro and millisecond timescale. To meet the challenge of analyzing the large, high-dimensional data sets generated by such simulations, advanced data mining tools (TRD2) will be implemented in VMD, including large-scale principal component analysis and k-medoids clustering.
The investigation of (1) and (2), as described above, will build on extensive previous work on both systems. For (S1), prior work includes the derivation of the first atomic model of the core structure of (S1) (Zhang) as well as studies on (S1)-associated sensory enzymes (Eisenbach) and bacterial flagella. For (S2), prior work includes studies on plant-degradation enzymes (Cann) as well as the flexibility and ultrastability (Gaub, Nash, Bayer) of (S2)-subcomponents. Extending these efforts, high-throughput modeling (TRD2) and MDFF techniques (TRD3), incorporating experimental data from biochemical assays (Bayer), AFM (Gaub, Nash), and EM (Zhang, Briegel), will be used to obtain complete structural models for (S1) and (S2). With high-resolution structures in hand, functionally critical areas of these complexes, e.g., the substrate and catalytic site of the (S1) and (S2) enzymes) will be treated using a hybrid QM/MM (TRD1) approach, enabling the study of ligand-binding and ATP-hydrolysis effects within (S1) as well as the effect of molecular-crowding on hydrolysis reactions in multiple sites within (S2). In NAMD, enhanced sampling (TRD1) techniques such as GSA will permit an investigation of long-timescale events in these multi-million atom complexes, including transmembrane signaling and receptor-mediated kinase activation within (S1) as well as highly-flexible cellulosomal linkers within (S2). TimeLine and parallel analysis (TRD2) tools will be used to identify and classify structural and dynamical features in the resulting high-dimensional trajectory data, permitting the quantitative characterization of the conformational dynamics of key functional proteins in their native molecular environments. Biochemical and genetic data will be employed to validate the structural models and simulation predictions obtained for (S1) (Parkinson, Eisenbach) and (S2) (Cann, Bayer).
