Emerging Studies
The biomedical sciences are enjoying a wealth of data from innovative new techniques in structure determination, DNA sequencing at the level of entire genomes, and direct manipulation and observation of single molecules. The Theoretical and Computational Biophysics Group complements these efforts through the most advanced molecular modeling, bioinformatic, and computational technologies, extending and refining these technologies in response to the experimental advances. Our research focuses on the modeling of large macromolecular systems in realistic environments. These efforts have produced insight into biomolecular processes coupled to mechanical force, bioelectronic processes in metabolism and vision, and the function and mechanism of membrane proteins. The most recent projects are listed below.
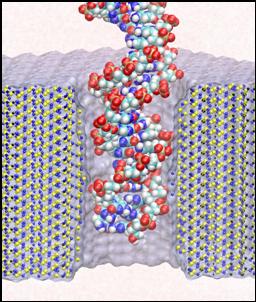
Nanoengineering
Each species from bacteria to human has a distinct genetic fingerprint. Therefore, detection of a single molecule of DNA represents the ultimate analytical tool for characterizing life forms with particular value in medical diagnostics and prevention of diseases. As a first step in the development of such a tool, a collaboration of electrical engineers and computational biologists has explored using a nanometer-diameter pore, manufactured into a nanometer-thick inorganic membrane, as a transducer that detects single molecules of DNA and produces an electrical signature of the structure. When an electric field is applied across the membrane, a DNA molecule immersed in electrolyte is attracted to the pore, blocks the current through it, and eventually translocates across the membrane as verified unequivocally by gel electrophoresis. The relationship between DNA translocation and blocking current has been established through molecular dynamics simulations. The simulations provide dynamic images of the nanopores as a kind of computational microscope, that images as well as analyzes the devices.
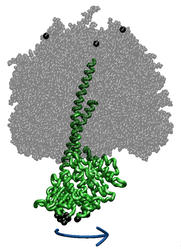
Torque Application to ATP Synthase Central Stalk
ATP synthase is a large multi-protein complex which includes a transmembrane Fo unit coupled to a solvent-exposed F1 unit via a central stalk. The stalk rotates within the surrounding subunits of F1, leading to cyclic conformational changes in the three catalytic sites in F1 and, thereby, to ATP synthesis. We use steered molecular dynamics to apply a torque to the central stalk in order to understand the cooperative interactions that underlie this mechanism.
Structure, Dynamics, and Function of Aquaporins
Aquaporins (AQPs) are a large family of membrane proteins facilitating the transport of water and small neutral soultes across the cell membrane. They are widely distributed in all life forms, including bacteria, plants, and mammals, and several pathological conditions in human are connected to impaired functions of these channels. Our extensive molecular dynamics simulations on human aquaporiun-1 (AQP1) and the E. coli glycerol facilitator (GlpF) have revealed several structural and functional features of these channels, including the highly conserved secondary structure elements, the mechanism and energetics of substrate permeation through the channel, and how they exclude protons while permitting fast water transport. See more about these channels on our Aquaporin main page.
Spectral Tuning in Rhodopsin Proteins
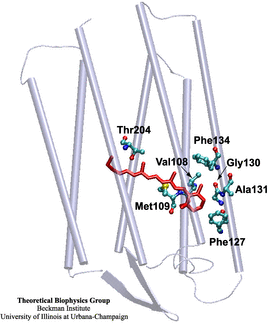
The rhodopsin receptors reside in the cell membrane, and function as sensors of light. These proteins consist of an apoprotein (opsin) and a retinal chromophore covalently bound to the apoprotein by a protonated Schiff base linkage to a lysine residue. While the protonated form of retinal Schiff base absorbs at about 440 nm in organic solvents, its maximal absorption is drastically changed after binding to the apoprotein, an effect known as 'opsin shift'. A fundamental challenge in vision research has been the elucidation of the physical mechanisms by which the protein matrix adjusts the maximal absorption of the chromophore, using the molecule retinal to detect light at different wavelengths. The spectral tuning in two very homologous rhodopsins, sensory rhodopsin II and bacteriorhodopsin, is investigated by means of a combined ab initio quantum mechanical/molecular mechanical calculation.
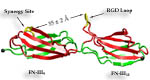
Fibronectin
Cells constitute only part of animal tissue, much of tissue is made of the extracellular matrix (ECM), a flexible mesh composed of several classes of macromolecule. Fibronectin (FN) is a component of the ECM that acts as a specific adhesive, forming networks that connect cells to the giant fibrous molecules that make up the majority of the ECM. Proper FN fibril formation is required to maintain prop er cell migration, thus FN plays roles in diseases affecting growth, development, tumors , and wound healing. We have used SMD simulations to examine how the mechanical force s present in the ECM affect FN fibril formation.
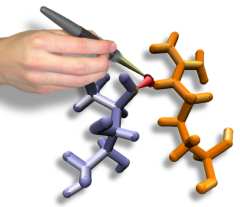
Interactive Molecular Dynamics and GlpF
Interactive Molecular Dynamics allows us to pull sugar molecules by hand through a simulation of the glycerol channel GlpF. As we push the virtual molecules around, we feel them in our hands as if they were real. We use this technique to explore features of the channel and gain new insights into the way it functions.
Molecular Dynamics Study of Rhodopsin
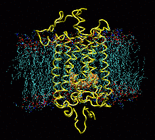
Responding to a great variety of ligands including hormones, neurotransmitters, and peptides, G-protein-coupled receptors (GPCRs) are the most important drug targets. Yet there is still little known about the first step of signal transduction. Rhodopsin is a member of this largest group of transmembrane receptors. The high resolution structure of rhodopsin determined by Palzcewski et al. (2000) gives us the opportunity to take a close look at the activation mechanism. Rhodopsin's ligand, retinal, which is covalently bound to the binding pocket, absorbs light at 500nm and undergoes a cis-trans isomerization triggering conformational changes in the protein. In order to understand the effects of the isomerization on the protein structure, we are performing molecular dynamics of a 40,000 atom model of rhodopsin in membrane.



