Steered Molecular Dynamics
Knowledge of the mechanism of association and dissociation of macromolecules is important for many biological structures and processes. Among the examples are the binding and dissociation of substrates of enzyme reactions, the recognition of ligands by their receptors or of DNA sequences by the DNA binding domains of regulatory proteins. These processes have in common a transition from one equilibrium state to another which often is a rare event on the time scale of molecular dynamics simulations of a few hundreds of picoseconds. Conventional computational methods for the sampling of barrier-crossing events increase the probability of unlikely configurations. Once such configurations have been sampled their actual occurence can be determined through the known relation between old and new probabilities. A methodologically related avenue to characterize rare events through molecular dynamics simulation is the addition of external forces which reduce the energy barriers. This approach has the advantage that it corresponds closely to micromanipulation through atomic force microscopy or optical tweezers.
The external force techniques can be applied to study many processes, including dissociation of avidin-biotin complex, dissociation of retinal from bacteriorhodopsin, stretching of DNA, etc.The forces can be defined using the MDScope graphics/MD software developed in our group
Avidin-Biotin Complex
We performed molecular dynamics simulations which induce, over periods of 40~ps to 500~ps, the unbinding of biotin from avidin by means of external harmonic forces with force constants close to those of AFM cantilevers. The applied forces are sufficiently large to reduce the overall binding energy enough to yield unbinding within the measurement time. Our study complements the work by Grubmueller et al. on biotin-streptavidin which employed a much larger harmonic force constant. The simulations reveal a variety of unbinding pathways, the role of key residues contributing to adhesion as well as the spatial range over which avidin binds biotin. In contrast to Grubmueller et al., the calculated rupture forces exceed by far those observed. We demonstrated, in the framework of models expressed in terms of one-dimensional Langevin equations with a schematic binding potential, the associated Smoluchowski equations, and the theory of first passage times, that picosecond to nanosecond simulation of ligand unbinding requires such strong forces that the resulting protein-ligand motion proceeds far from the thermally activated regime of millisecond AFM experiments, and that simulated unbinding cannot be readily extrapolated to the experimentally observed rupture.
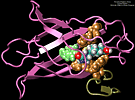 MD simulations of protein-ligand adhesion for the avidin-biotin complex.
MD simulations of protein-ligand adhesion for the avidin-biotin complex.
PLA2
We are applying the steered molecular dynamics method to investigate the action of human synovial protein phospholipase A2 (PLA2) at the lipid water interface. PLA2 binds to a membrane surface in a scooting mode and catalyzes the hydrolysis reaction of the sn-2 ester bond of phospholipids which subsequently results in the destruction of membrane surface. The actual catalysis is believed to be initiated through the suction of a phospholipid from the membrane surface into the hydrophobic channel of PLA2 where the reaction takes place. The mechanism of this suction is little understood and is hypothesized to be caused by destabilization of the polar phospholipid headgroup by the hydrophobic environment of the binding channel of PLA2. We investigate the process of extrusion of the phospholipid for two modes of coupling of PLA2 to the membrane surface, i. e., tightly and loosely bound modes. Our hypothesis is that prior to extruding the phospholipid, PLA2 must form the tightly bound complex, while the loosely bound complex should not lead to catalysis.
Retinal's Binding Pathway in bR
Formation of bacteriorhodopsin (bR) from the apoprotein and retinal has been studied experimentally, but the actual pathway, including the site of retinal entry, is little understood. Molecular dynamics simulations provide a surprisingly clear prediction. A window between bR helices E and F in the trans-membrane part of the protein can be identified as an entry site for retinal (purple van der Waals spheres in this figure). Steered molecular dynamics, performed by applying a series of external forces of 200--1000 pN over a period of 0.2 ns to retinal, allow one to extract retinal from bR once the Schiff base bond to Lys216 (blue van der Waals spheres) is cleaved. Extraction proceeds until the retinal tail forms a hydrogen bond network with side groups lining the entry window. The manipulation induces a distortion with a fitted RMS deviation of coordinates (ignoring retinal, water, and hydrogen atoms) of less than 1.9 Angstroms by the time the entry window--retinal tail hydrogen bond network forms. The forces used are consistent with those expected for fast extraction and do not indicate significant potential barriers. We suggest that the path followed in the simulated extraction of retinal also describes the binding of retinal. The simulation reveals that movement of a water molecule between Lys216 and retinal's aldehyde group can initiate unbinding.
References





