C&S7: DNA Membrane Channels
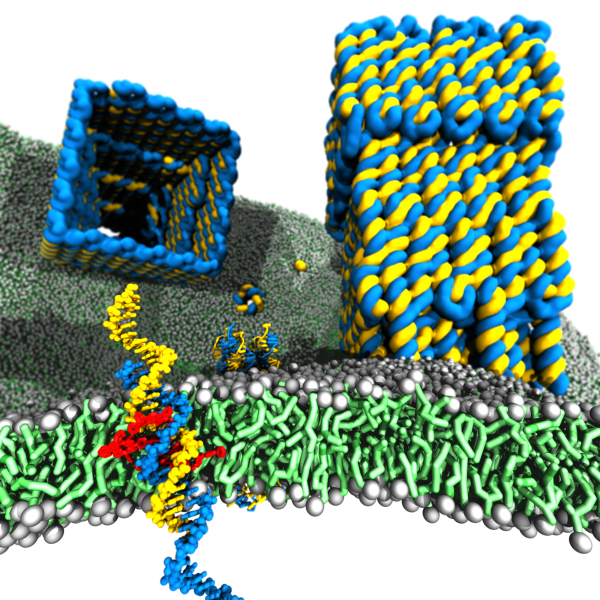
Membrane protein channels involved in cellular signal transductions are fascinating biological sensors with high selectivity and efficiency. Recently, DNA origami nanostructures emerged as highly customizable mimetics of biological membrane channels. A typical DNA membrane channel is assembled from several DNA double helices arranged into a polygon pattern, with the central cavity forming the transmembrane pore. To facilitate insertion of the DNA channel into a lipid bilayer membrane, the DNA helices are chemically modified to carry hydrophobic anchors. Until now, most of the DNA channels featured four or six DNA helices arranged in a square or a hexagon, with the inner channel diameter varying between 1 and 2.5 nm. The outstanding problems of the field are: (1) Establishing the relationship between the design of a DNA channel and its function as a passage across the lipid bilayer membrane. (2) Improving the designs of DNA channels to realize selective transmembrane transport controlled by external stimuli. (3) Engineering DNA channels for applications in sensing, drug delivery and artificial tissues technologies.
Experimental characterization of DNA origami is essential for accurate design, but has been limited to rather qualitative experimental techniques such as atomic force spectroscopy, small-angle X-ray scattering, and transmission electron microscopy (TEM). The only atomic-level model of DNA origami in situ has been derived from cryo-electron microscopy (cryo-EM), which revealed considerable deviations from the idealized design. Predictive computational modeling of DNA origami objects is an attractive alternative to experimental characterization procedures, which are expensive and time-consuming. Currently, the most accurate computational method that can address the outstanding problems of the field is the all-atom molecular dynamics method. Due to their relatively large dimensions (of 30 nm on each side or larger), using the Center-developed software, NAMD (TRD1), is essential to make modeling of DNA origami channels feasible.
In 2013, we have reported the first all-atom molecular dynamics simulation of DNA origami objects. In collaboration with the Keyser group, we identified the key factors affecting the ionic conductivity of a DNA origami plate. The Keyser group demonstrated that DNA channels made of four or six helices can be embedded into a bilayer membrane with the help of two covalently-labeled cholesterol or porphyrin groups, although the ionic conductance of such DNA channels was found to vary over a considerable range, suggesting multiple conductance states of the channel. Last year, we reported the first all-atom molecular dynamics simulation of a 6-helix DNA channel. Our MD simulations showed that, while overall remaining stable, the local structure of the channel undergoes considerable fluctuations, departing from its idealized design. Furthermore, we found the ionic conductance of the DNA channel to depend on the membrane tension, making them potentially suitable for force sensing applications.
In close collaboration with the Keyser group, we will continue to explore DNA channels of different architectures. Our all-atom MD simulations will characterize the conductance mechanism of improved DNA channels at atomic resolution and predict the effect of functional groups on the selectivity of the channels. As the first step toward expanding the design space of the channels, we have just demonstrated a DNA channel of low ionic conductance, consisting of just one DNA helix, and a large porin-like structure, consisting of 112 DNA helices, that has a conductance similar to that of the nuclear pore complex. We have also begun to explore structural deformation as a mechanism of voltage gating and incorporation of unusual DNA motives to induce selective transport. A unique aspect of this collaboration is that the electric current recordings of the Keyser group can be matched one-to-one with the simulated ionic conductance, making validation of structural models and their rational design based on computations feasible.



