C&S2: Molecular Mechanism of Drug Interactions in Cytochrome P450
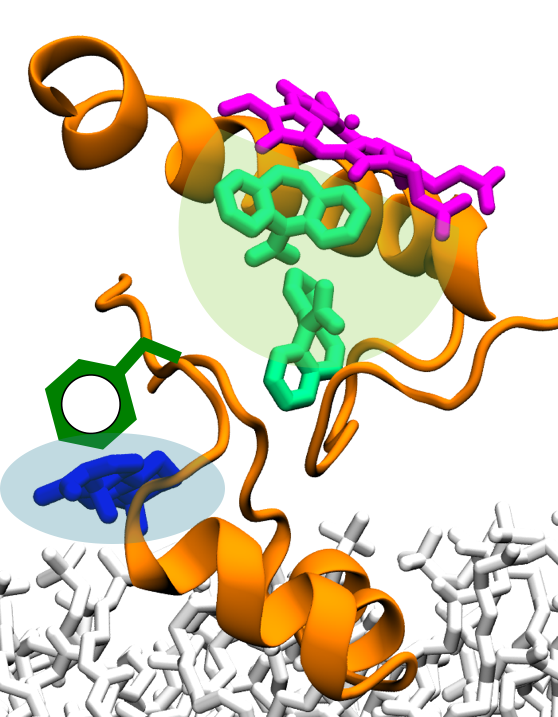
Adverse drug-drug interactions (DDI) are a significant problem in clinical practice, especially for patients taking multiple drugs on a regular basis. A major type of DDI involves the interference of different drugs at the level of metabolism, and is observed when the metabolism of one drug is affected by the presence of co-administered drugs. In the human body, 70% of drugs are metabolized by cytochromes P450 (CYPs), a large family of enzymes involved in the biotransformation of drugs. Hence, drug metabolism mediated by CYPs constitutes a major determinant in the occurrence of several adverse DDI. Therefore, a detailed description of the molecular mechanism underlying DDI mediated by CYPs will provide medicinal chemistry and pharmacology with a way to prevent adverse drug effects in clinical practice.
In the human body, CYPs are membrane-associated proteins. A large body of evidence suggests that drug recruitment by CYPs is determined by their partition and orientation in the membrane. Therefore, to gain understanding of the drug binding mechanism and DDI in CYPs, it is crucial to characterize their structure and dynamics in the membrane-associated form. Current experimental techniques, including FRET and linear dichroism, provide a valuable yet limited description of the insertion and orientation of CYPs in membrane-like environments. Thus, to understand the interaction of CYPs with the membrane at an atomic resolution, MD simulations performed with Center-developed software NAMD (TRD1) are employed to generated atomic-level models of membrane-associated CYP enzymes. Furthermore, in addition to reliable structural models of CYPs, succesful characterization of DDI relies on adequate simulation parameters of multiple drugs. For this purpose, the force field Toolkit developed in the Center (TRD2) will be employed for parametrization of small molecules.
A detailed description of the molecular mechanism of DDI mediated by CYPs requires a concerted effort involving molecular models and simulations, as well as experiments. Recently, the Center has derived the membrane-bound form of CYP3A4 (Sligar), one of the major drug-metabolizing enzymes in the human body. The model derived from simulations was validated experimentally, employing linear dichroism to measure the orientation of the enzyme in Nanodiscs, yielding excellent agreement between simulations and experiments. The resulting membrane-associated form of CYP3A4 has been recently employed to initially probe the mechanism of DDI mediated by membrane-bound CYP3A4 with atomic detail (Sligar). Moreover, the Center has succesfully employed the general simulation approach to model membrane association of CYP enzymes to CYP2J2 (Das), another human CYP. The derived CYP2J2 model is currently being employed to study the mechanism of drug binding for different small molecules.
An innovative aspect of the proposed work is that, for the first time, the characterization of DDI in CYPs will be done in the explicit presence of the membrane, combining membrane-bound MD simulations and experiments performed in Nanodiscs. An ensemble of protein structures in the presence of the membrane obtained from MD simulations will be employed to perform massive docking of drugs, to take into account con- formational changes of the protein in its native membrane environment that might be relevant for drug binding. Clustering analysis of the generated enzyme-drug complexes will be performed with the analysis tools included in VMD (TRD2). This aspect will be complemented with a computational study of drug membrane partitioning, emplyoying bias-exchange umbrella sampling as implemented in NAMD (TRD1). Adequate simulation parameters for the studied drugs will be generated employing the forcefield Toolkit (TRD2).



