- The Chain-like Elasticity of Titin -
Titin, a Molecular Spring
The ability of striated muscle to stretch and relax,
in an elastic
manner, is crucial for its role as a force bearing component in higher
organisms. An individual muscular contractile unit, called the
muscle sarcomere, extends beyond its equilibrium length immediately
following contraction, and a passive restoring force arises from a
protein called titin, whose elastic response restores a muscle fiber to
its resting length. Titin is the largest known protein, with a
length greater than 1 µm and a molecular weight of up to 4
MDa. It is built from a modular construct of ~300 tandem repeats
of predominantly immunoglobulin-like (Ig) and fibronectin type III
(FnIII) domains, and from flexible random coil-like PEVK (rich in
proline, glutamate, valine, and lysine) regions; there exist
furthermore single Ig-domain insertions that distinguish isoforms of
titin in cardiac and skeletal muscle. The N-terminal section of
titin originates from the sarcomeric Z-disc and its C-terminal region
is an integral component of the sarcomeric M-line
Figure 1 depicts a schematic view of titin's individual
components, and its position within the muscle sarcomere.

Figure 1: Schematic of the
titin spring in muscle, and
detailed view of titin's Z1Z2 domains. Shown is an electron
micrograph image (courtesy R. Craig, University of Massachusetts, USA)
of the human muscle sarcomere
and the schematic representation of titin
along the sarcomere length. Titin is anchored with one of its
ends (N-terminal) through the protein telethonin to the sarcomeric
Z-disc and with its other end (C-terminal) to the M-line. Components of
titin shown in this figure include the proximal
Z1Z2 domains (circled),
entropic PEVK region, the well studied I27 (now numbered I91) domain in
the I-band, and the Ig-FnIII triad A168-170 of the A-band.
Extensive studies have characterized titin as a protein with many
distinct functions. At the early stages of myofibril growth,
titin is believed to play a crucial role as a molecular scaffold for
organizing the various components of the sarcomere, as evidenced by
exact matches between the pattern of repeat domains in titin and myosin
(thick filament) periodicity. There is also evidence that the
ends of titin, not only anchor at, but interact with signalling
proteins at the Z-line and M-line, possibly mediated by passive
stretching forces. The very N-terminal region of titin relies on
the binding of Z1 and Z2 to the ligand telethonin, one of the most
abundant transcripts in striated muscle. Binding studies have
revealed that it is highly specific for both titin Z1 and Z2, but not
for Z1 or Z2 individually. Biochemical studies of telethonin have
shown that its presence, in conjunction with titin Z1Z2, is required
for progression of muscle growth. Point mutations to telethonin
resulting in premature stop codons have been correlated to a form of
limb girdle muscular dystrophy, suggesting that telethonin plays a
major role in the stabilization of N-terminal titin at the
Z-disc. Recently the structure of titin Z1Z2, both bound to
telethonin, and separately crystalized have been solved. A
subsequent investigation involving molecular
dynamics studies of the titin Z1Z2-telethonin complex
revealed that telethonin is in fact a component of the structure that
anchors the N-terminal region of titin to the sarcomere.
Here we turn our attention to the unbound structure for titin Z1Z2 is
the first tandem Ig-domain for titin ever to be resolved, which
provides a means to study whether interactions between connected
domains (i.e. interdomain forces, which to this point have not been
significantly investigated) play a significant role in the overall
elasticity of titin.
Secondary and Tertiary Structure Elasticity of Titin Z1Z2 and a Titin Chain Model
Investigations of the extensibility and elasticity of titin, both \textit{in situ} and at the level of single molecules, have revealed that titin is not merely a soft entropic spring, but rather exhibits also a stiffer, structure-dependent elasticity. Fig.~\ref{fig:FIG8} shows a schematic presentation of titin elasticity across a wide range of extension and force as studied by optical tweezers and atomic force microscopy (AFM). This figure illustrates that a region of soft elasticity at weak forces ($<$50~pN) is followed by a region of non-linear stiff elasticity at forces of $>$50~pN. While the stretching and unfolding of individual or linked PEVK, Ig-, and FN-III domains connected with the soft and stiff elasticity of titin have been extensively characterized through AFM, optical tweezers, and computational unfolding studies, an elasticity of titin associated with domain-domain extension, has been less extensively investigated.

Figure 2: Two regimes of titin elasticity - soft, and stiff non-linear. Shown schematically is how titin responds to stretching forces of various magnitudes.. At weak forces (<50 pN), the primary contribution of titin's response arises from the PEVK domains and the straightening of a bent and twisted titin chain. At high forces (>50 pN) titin exhibits a highly non-linear elastic response. In this study we will relate the non-PEVK soft elasticity of titin to the so-called tertiary structure elasticity and the non-linear stiff elasticity to the secondary structure elasticity.
The soft elasticity of titin is likely due, in part, to a generic behavior of random coils formed by PEVK and other similar domains, an entropic spring behavior in principle well understood. The studies of stiff elasticity were based on the then available structures of titin domains I1 and I91 (formerly I27) and revealed that the individual domains, when subject to sufficiently strong stretching forces, unravel their secondary structure, giving rise to titin's non-linear stiff elasticity called secondary structure elasticity. The soft elasticity of titin is also due to a straightening out of titin's multi-domain segments. Adjacent segments may exhibit a flexibility that leads a titin chain to assume in the relaxed state a locally compacted shape, and a straight shape when under tension. The question arises if the elasticity linked to such structural transition contributes to the soft elasticity of titin characterized through a spring constant that falls in a range of observed values.
Lacking until recently structures of two or more adjacent titin domains made it impossible to computationally investigate titin's soft elasticity associated with domain-domain rearrangement. The relative orientation of adjacent domains should be controlled through the respective linker as well as domain-domain interaction. The tertiary structure specifying the arrangement of adjacent domains should resist forces that stretch domain pairs apart, thereby giving rise to a local elastic property, to be referred in this paper as tertiary structure elasticity. Contributions of many adjacent domain pairs can add and confer to titin an overall elasticity. Fortunately, the structure of a tandem of two adjacent Ig-domains, Z1Z2 and three consecutive domains A168-170 have been recently elucidated by X-ray crystallography. In addition, data on the conformation and dynamics of Z1Z2 in solution have become available through small angle X-ray scattering (SAXS) and NMR spectroscopy. These data allow us now to explore the extent to which the dynamics of interdomain arrangement contributes to the elasticity of the titin chain.
Given the wealth of experimental data on Z1Z2, our study will use this tandem as a model system to explore the role of interdomain relations in titin elasticity. Crystallographic data revealed Z1Z2 in two conformational states, one in an overall ``V''-shape shown in Figure 3A and one in an extended shape shown in Figure 3B. Z1Z2 models satisfying NMR residual dipolar coupling (RDC) data and SAXS measurements in solution are shown in Figure 3C. The models correspond to a structure that is intermediate between those shown in Figures 3A and B. The three structures showing Z1Z2 in closed, intermediate, and wide open ``V''-shape suggest immediately that interdomain dynamics might contribute to the tertiary structure elasticity of titin. A highly schematic representation of this elasticity is shown in Figure 3D, in which the tandem domains become extended while preserving secondary structure integrity of titin's domains. However, it is yet to be established whether the multiple conformations accessible to Ig tandems in the filament are, or not, energetically equivalent and how they contribute to elasticity in titin. This information is best obtained by means of molecular dynamics simulations.
 Figure 3:
Experimentally derived conformers for titin Z1Z2
and a possible mode of tertiary structure elasticity. (A) and (B)
show the crystal structure (PDB code 2A38) for the N-terminal region of
titin comprised of the tandem Z1 (purple) and Z2 (orange) domains in
two crystallographically constrained conformations, termed closed, and open, respectively. (C)
NMR-RDC models of the experimentally observed semi-extended
conformation for Z1Z2. The colors correspond to the NMR lowest
(blue) and second lowest (red) energy conformers and the small angle
X-ray scattering best fit conformer (green). (D) Highly
schematic view of the tertiary structure elasticity of titin due to
bending adjacent protein domains open and closed. In actual
titin, not every linker may contribute high flexibility; some linkers
might be short and stiff.
Figure 3:
Experimentally derived conformers for titin Z1Z2
and a possible mode of tertiary structure elasticity. (A) and (B)
show the crystal structure (PDB code 2A38) for the N-terminal region of
titin comprised of the tandem Z1 (purple) and Z2 (orange) domains in
two crystallographically constrained conformations, termed closed, and open, respectively. (C)
NMR-RDC models of the experimentally observed semi-extended
conformation for Z1Z2. The colors correspond to the NMR lowest
(blue) and second lowest (red) energy conformers and the small angle
X-ray scattering best fit conformer (green). (D) Highly
schematic view of the tertiary structure elasticity of titin due to
bending adjacent protein domains open and closed. In actual
titin, not every linker may contribute high flexibility; some linkers
might be short and stiff. Accordingly, we employed MD simulations to study the conformational dynamics of the titin Z1Z2 tandem, focusing specifically on the forces governing domain-domain interactions and associated elastic properties. Extended timescale simulation of the open Z1Z2 conformer reveal, independently, a quasi-equilibrium structure that matches precisely the experimentally observed semi-extended NMR-RDC conformer. Using a combination of steered molecular dynamics (SMD) and adaptive biasing force (ABF) simulations, we calculate the work required to hinge apart the Z1 and Z2 domains without unfolding either Ig-domain, thereby exploring only the contribution from tertiary structure characteristics. We then extend our scope by using the simulation results to construct a schematic stochastic model for a system of titin Ig-domain repeats (an Ig-chain), in which changes in tertiary structure endows the chain with overall elasticity.
Solving the Solution Conformation of Titin Z1Z2
Previously reported NMR-RDC conformers for Z1Z2 suggested that the preferential conformation of Z1Z2 in solution is, in fact, an intermediate state between the open and closed forms observed in the crystal structure, termed the semi-extended state. Indeed, during simulation Z1Z2 approached this semi-extended state as clearly revealed in a superposition of the NMR-RDC structure and the MD trajectory, shown in Figure 4C.
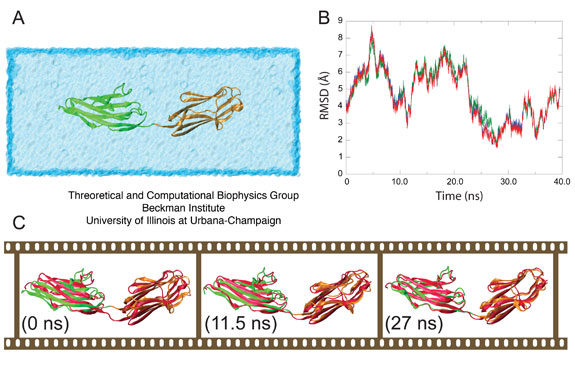
The root mean square deviation (RMSD) between (i) the backbone α-carbons of the semi-extended secondary structural elements of the MD model, (ii) the lowest energy NMR-RDC conformer, (iii) the second lowest energy NMR-RDC conformer, and (iv) the RDC conformer best fitting SAXS data was calculated in order to assess the structural fit between the computational model and the molecule in solution. Models (ii -- iv) represent the experimentally observed conformation of Z1Z2 in solution. Although all fits were comparable, the closest agreement was found with model (iii). The result (Figure 3B) reveals that the fit is achieved after 22 ns of simulation, at which point the two ends of Z1Z2 begin to hinge towards each other, achieving remarkable structural alignment with the mentioned NMR conformer as shown in Figure 3C, illustrating the gradual bending of Z1Z2 from its open state to the semi-extended state. The MD simulations were performed by us before the NMR structures became available, and the convergence of both the MD model, in which the semi-extended state for Z1Z2 remained stable for ~10 ns, and the observed NMR conformers suggests strongly that the energetically viable semi-extended conformation reported here represents a significant quasi-equilibrium state for solvated tandem Z1Z2 Ig-domains.
Tertiary Structure Elasticity of Titin Z1Z2
In order to characterize this type of elasticity, we ask what force is required to alter the arrangement of tandem Ig-domains, i.e., to stretch the elements of the chain out by reorienting neighboring domains, but without unfolding the domains. For this purpose, a series of low force SMD simulations were performed on the closed Z1Z2 conformer, in which Z1 was held fixed and Z2 permitted to rotate about the linker region, to `open' the tandem complex. A force of 50 pN applied to the C-terminus of Z2 over 2 ns was found not to be sufficient to open Z1Z2; however, forces of 94 pN and 164.5 pN opened the pair within 2 ns without unfolding either domain. This suggests that bending at the Z1Z2 hinge is possible with rather small forces (comparatively, at MD timescales). It is desirable, then, to determine the free energy profile that governs the mechanical domain-domain behavior.
Before continuing, we note that forces mentioned, i.e., 90-160 pN, are stronger than forces known to extend titin chains using the most sensitive single molecule force spectroscopy techniques such as optical tweezers. We will show here that for a chain of many domains, forces on the order of 50 pN, which are comparable to those employed with optical tweezers, can stretch chains by ~600 Å; this is possible since every individual domain is stretched by a few Å, while the 90-160 pN forces lead to larger stretching and bending, e.g., stretching of 40 Å, in one domain pair and of 1.2 µm for a 300 domain chain. Naturally the effect of strong forces is relatively easy to discern in simulations. To describe the effect of weak forces is relatively difficult and requires the calculation of the potential of mean force (free energy profile) linked to domain-domain bending.
The calculation, by means of MD simulations, of a free energy profile for molecular motions, such as opening Z1Z2 around its centered hinge, requires significant sampling. While sampling through multiple SMD simulations to reconstruct the potential of mean force (PMF) along a reaction coordinate has been previously achieved for systems involving conduction of small molecules through membrane channels, this method has not been employed for either the present system or similar systems. Whereas the potential of mean force linked to stretch an α-helix in vacuum has been successfully determined by means of SMD, an analogous application to solvated helices presently is beyond the reach of the SMD approach. However, an application of the ABF method to stretch a solvated helix and determine the corresponding PMF has been successful. We employ, therefore, the ABF method.
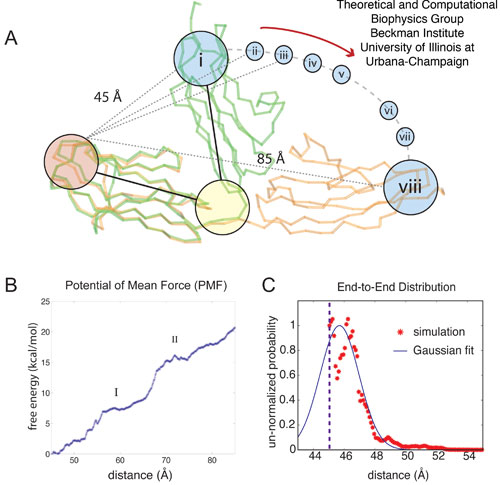
Figure 5: Adaptive biasing force (ABF) simulations of Z1Z2. The ABF simulations of titin Z1Z2 are depicted schematically in (A); in the simulations, the crystallographic structure for the closed form of Z1Z2 (green) is hinged towards the open conformation and the potential of mean force (PMF) profile for this motion is calculated. The ABF simulations step through the states (i - viii), each state stepping through an arc motion of 5 Å, (including one 10 Å, window). The PMF profile over the extension range 45-85 Å, i.e., for transitioning between the closed and open state, is plotted in (B), with I and II denoting intermediate states along the trajectory describing the stretched Z1Z2. The trough at II corresponds to the semi-extended conformer for Z1Z2 observed both in experiment and simulation. The distribution defined through Eq. 33, is shown in (C) (red stars) along with the Gaussian distribution defined in Eq. 44 (blue line). The dashed line at 45 Å, denotes the beginning of the simulation range.
Uncovering an Unfolding Intermediate State for Titin Z2, but not Z1
Finally, we study also the non-linear stiff elasticity of titin. This elasticity is due to the interactions within the secondary structure elements of titin, namely Ig- and FnIII-like domains which strongly resist mechanical stretching forces and have been well characterized in the studies mentioned above. In this case, we probe by means of simulations the secondary structure elasticity of individual titin Z1 and Z2 domains, as done earlier for I1 and I91.
Our findings reveal that although Z1 and Z2 share similar architecture and fold, when stretched at low forces (relatively, for simulations at 350~pN) Z1 unfolds differently from Z2, with Z2 exhibiting the stepped unfolding intermediate seen also classically in titin I27 (now I91). Z1 however, unfolded without pausing briefly at this transition state. This surprising finding was correlated with a difference in the composition of the force bearing terminal Β-strand of Z1 (which has a sequence VAL-VAL-VAL) and Z2 (which has a sequence of MET-THR-VAL; this suggests that the irregular peptide sequence at the Z2 A' strand likely reinforces its interactions with the opposing G Β-strand, leading to its characteristic delay in unfolding. The search for stable intermediate unfolding conformations of mechanical proteins, therefore, may benefit from similar sequence comparison along their force bearing components as a means for predicting the response to stretching forces. These results are shown in Figure 6.
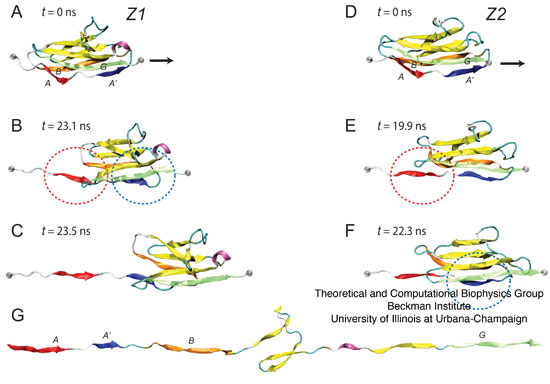
Figure 6: Stretching individual Igdomains of Z1 and Z2. (A–C) Snapshots of the unfolding trajectory for titin Z1 under 350 pN constant force: (A) at t = 0 ns; (B) at t = 23.1 ns, i.e., the moment of simultaneous detachment of A and A' strands (denoted by red and blue circles corresponding also to the strand color); and (C) at t = 23.5 ns, i.e., when the N-terminal region of Z1 extends. (D,E) Snapshots of the unfolding trajectory for titin Z2 under 350 pN constant force: (D) at t = 0 ns; (E) at t; 19.9 ns when strand A detaches (red circle); and (F) at t ~ 22.3 ns when strand A9 detaches (blue circle). An unfolding intermediate, in which the A'G strand holds briefly for ~3 ns (see also Fig. 9 B) after the AB strand detaches, is seen during this unraveling of Z2, but not during the unraveling of Z1. After steps C and F, both Z1 and Z2 unfold quickly with little resistance, toward the elongated form shown in panel G.
- Click here for a movie (mpeg, 4.5M) of the
equilibration trajectory in which titin Z1Z2 transitions from the
extended to the semi-extended state. The simulated Z1Z2 system is shown
in green and orange, and is
superimposed with the second lowest energy NMR conformer for Z1Z2 reported above. - Click here for a movie (mpeg, 3.1M) showing an unfolding trajectory in which titin Z1 is stretched with a 350 pN force.
- Click here for a movie (mpeg, 2.6M) showing an unfolding trajectory in which titin Z2 is stretched with a 350 pN force
Publications and Related Commentary
Secondary and tertiary structure elasticity of titin Z1Z2 and a titin chain model. Eric H. Lee, Jen Hsin, Olga Mayans, and Klaus Schulten. Biophysical Journal, 93:1719-1735, 2007.Molecular mechanisms of cellular mechanics. Mu Gao, Marcos Sotomayor, Elizabeth Villa, Eric Lee, and Klaus Schulten. Physical Chemistry - Chemical Physics, 8:3692-3706, 2006.
Mechanical strength of the titin Z1Z2/telethonin complex. Eric H. Lee, Mu Gao, Nikos Pinotsis, Matthias Wilmanns, and Klaus Schulten. Structure, 14:497-509, 2006.
Also see a commentary
of our work in the journal Structure.
Investigators
- Eric H. Lee
- Jen
Hsin
- Klaus Schulten
- Olga Mayans (University of Basel)
Related TCB Group Projects
Page created and maintained by Eric H. Lee.
