



Next: Defining and setting up
Up: Collective Variable-based Calculations (Colvars)1
Previous: A crash course
Contents
Index
Subsections
Here, we document the syntax of the commands and parameters used to set up and use the Colvars module in NAMD.
One of these parameters is the configuration file or the configuration text for the module itself, whose syntax is described in ![[*]](crossref.png) and in the following sections.
and in the following sections.
<18135>>
The Colvars module is accessed in VMD through the command cv.
The command must be used the first time as cv molid  molid
molid to set up the Colvars module for a given molecule.
In all following uses, the cv command will continue operating on the same molecule, regardless of its ``top'' status.
To use the cv command on a different molecule, use cv delete first and then cv molid
to set up the Colvars module for a given molecule.
In all following uses, the cv command will continue operating on the same molecule, regardless of its ``top'' status.
To use the cv command on a different molecule, use cv delete first and then cv molid  molid
molid .
Invoking the cv command with no arguments prints a help screen.
.
Invoking the cv command with no arguments prints a help screen.
<18136>>
Collective variables and biases can be added, queried and deleted through the scripting command cv, with the following syntax: cv  subcommand
subcommand [args...].
For example, to query the value of a collective variable named myVar,
use the following syntax: set value [cv colvar myVar value].
All subcommands of cv are documented below.
[args...].
For example, to query the value of a collective variable named myVar,
use the following syntax: set value [cv colvar myVar value].
All subcommands of cv are documented below.
2]
- configfile
 file name
file name : read configuration from a file;
: read configuration from a file;
- config
 string
string : read configuration from the given string; both config and configfile subcommands may be invoked multiple times;
: read configuration from the given string; both config and configfile subcommands may be invoked multiple times;
- reset: delete all internal configuration of the Colvars module;
- version: return the version of the colvars code.
- list: return a list of all currently defined variables;
- list biases: return a list of all currently defined biases (i.e. sampling and analysis algorithms);
- load
 file name
file name : load a collective variables state file, typically produced during a previous simulation;
: load a collective variables state file, typically produced during a previous simulation;
- load
 prefix
prefix : same as above, but without the .colvars.state suffix;
: same as above, but without the .colvars.state suffix;
- save
 prefix
prefix : save the current state in a file whose name begins with the given argument; if any of the biases have additional output files defined, those are saved as well using the same prefix;
: save the current state in a file whose name begins with the given argument; if any of the biases have additional output files defined, those are saved as well using the same prefix;
- update: recalculate all colvars and biases based on the current atomic coordinates;
- addenergy
 E
E : add value E to the total bias energy; this must be used within calc_colvar_forces;
: add value E to the total bias energy; this must be used within calc_colvar_forces;
- printframe: return a summary of the current frame, in a format equivalent to a line of the collective variables trajectory file;
- printframelabels: return text labels for the columns of printframe's output;
- bias
 name
name energy: return the current energy of the bias
energy: return the current energy of the bias  name
name ;
;
- bias
 name
name update: recalculate the bias
update: recalculate the bias  name
name ;
;
- bias
 name
name delete: delete the bias
delete: delete the bias  name
name ;
;
- bias
 name
name getconfig: return config string of bias
getconfig: return config string of bias  name
name .
.
<18402>>Collective variables and biases can be added, queried and deleted through the scripting command cv, with the following syntax: cv  subcommand
subcommand [args...].
For example, to query the value of a collective variable named myVar,
use the following syntax: set value [cv colvar myVar value].
All subcommands of cv are documented below.
[args...].
For example, to query the value of a collective variable named myVar,
use the following syntax: set value [cv colvar myVar value].
All subcommands of cv are documented below.
<22332>>
- configfile
 file name
file name : read configuration from a file;
: read configuration from a file;
- config
 string
string : read configuration from the given string; both config and configfile subcommands may be invoked multiple times;
: read configuration from the given string; both config and configfile subcommands may be invoked multiple times;
- reset: delete all internal configuration of the colvars module;
- version: return the version of the colvars code.
- list: return a list of all currently defined variables;
- list biases: return a list of all currently defined biases (i.e. sampling and analysis algorithms);
- load
 file name
file name : load a collective variables state file, typically produced during a simulation;
: load a collective variables state file, typically produced during a simulation;
- save
 prefix
prefix : save the current state in a file whose name begins with the given argument; if any of the biases have additional output files defined, those are saved as well;
: save the current state in a file whose name begins with the given argument; if any of the biases have additional output files defined, those are saved as well;
- update: recalculate all colvars and biases based on the current atomic coordinates;
- printframe: return a summary of the current frame, in a format equivalent to a line of the collective variables trajectory file;
- printframelabels: return text labels for the columns of printframe's output;
- colvar
 name
name value: return the current value of colvar
value: return the current value of colvar  name
name ;
;
- colvar
 name
name update: recalculate colvar
update: recalculate colvar  name
name ;
;
- colvar
 name
name type: return the type of colvar
type: return the type of colvar  name
name ;
;
- colvar
 name
name delete: delete colvar
delete: delete colvar  name
name ;
;
- colvar
 name
name addforce
addforce  F
F : apply given force on colvar
: apply given force on colvar  name
name ;
;
- colvar
 name
name getconfig: return config string of colvar
getconfig: return config string of colvar  name
name .
.
- colvar
 name
name cvcflags
cvcflags  flags
flags : for a colvar with several cvcs (numbered according to their name
string order), set which cvcs are enabled or disabled in subsequent evaluations according to a list of 0/1 flags (one per cvc).
: for a colvar with several cvcs (numbered according to their name
string order), set which cvcs are enabled or disabled in subsequent evaluations according to a list of 0/1 flags (one per cvc).
- bias
 name
name energy: return the current energy of the bias
energy: return the current energy of the bias  name
name ;
;
- bias
 name
name update: recalculate the bias
update: recalculate the bias  name
name ;
;
- bias
 name
name delete: delete the bias
delete: delete the bias  name
name ;
;
- bias
 name
name getconfig: return config string of bias
getconfig: return config string of bias  name
name .
.
To enable a Colvars-based calculation, two parameters must be added to the NAMD configuration file, colvars and colvarsConfig.
An optional third parameter, colvarsInput, can be used to continue a previous simulation.
- colvars
 Enable the Colvars module
Enable the Colvars module 
Context: NAMD configuration file
Acceptable Values: boolean
Default Value: off
Description: If this flag is on, the Colvars module within
NAMD is enabled; the module requires a separate configuration
file, to be provided with colvarsConfig.
- colvarsConfig
 Configuration file for the collective variables
Configuration file for the collective variables 
Context: NAMD configuration file
Acceptable Values: UNIX filename
Description: This file contains the definition of all collective variables and
their biasing or analysis methods.
This file can also be provided by the Tcl command cv configfile; alternatively, the contents of the file itself can be given as an argument to the command cv config.
- colvarsInput
 Input state file for the collective variables
Input state file for the collective variables 
Context: NAMD configuration file
Acceptable Values: UNIX filename
Description: When continuing a previous simulation run, this file contains the current state of all collective variables and of their associated algorithms.
It is written automatically at the end of any simulation with collective variables.
This file can also be provided by the Tcl command cv load.
All the parameters defining the colvars and their biasing or analysis algorithms are read from the file specified by the configuration option colvarsConfig, or by the Tcl commands cv config and cv configfile.
Hence, none of the keywords described in this section and the following ones are available as keywords for the
NAMD configuration file.
The syntax of the Colvars configuration is ``keyword value'', where the keyword and its value are separated by any white space.
The following rules apply:
- keywords are case-insensitive (upperBoundary is the same as upperboundary and UPPERBOUNDARY): their string values are however case-sensitive (e.g. file names);
- a long value or a list of multiple values can be distributed across multiple lines by using curly braces, ``{'' and ``}'': the opening brace ``{'' must occur on the same line as the keyword, following a space character or other white space; the closing brace ``}'' can be at any position after that;
- many keywords are nested, and are only meaningful within a specific context: for every keyword documented in the following, the ``parent'' keyword that defines such context is also indicated in parentheses;
- the `=' sign between a keyword and its value, deprecated in the NAMD main configuration file, is not allowed;
- Tcl syntax is generally not available, but it is possible to use Tcl variables or bracket expansion of commands within a configuration string, when this is passed via the command cv config ...; this is particularly useful when combined with parameter introspection (see 2.2.2), e.g. cv config "colvarsTrajFrequency [DCDFreq]";
- if a keyword requiring a boolean value (yes|on|true or no|off|false) is provided without an explicit value, it defaults to `yes|on|true'; for example, `outputAppliedForce' may be used as shorthand for `outputAppliedForce on';
- the hash character # indicates a comment: all text in the same line following this character will be ignored.
The following keywords are available in the global context of the Colvars configuration, i.e. they are not nested inside other keywords:
To illustrate the flexibility of the Colvars module, a non-trivial setup is represented in Figure 6.
The corresponding configuration is given below. The options within the colvar blocks are described in ![[*]](crossref.png) and
and ![[*]](crossref.png) , those within the harmonic and histogram blocks in
, those within the harmonic and histogram blocks in ![[*]](crossref.png) .
Note: except colvar, none of the keywords shown is mandatory.
.
Note: except colvar, none of the keywords shown is mandatory.
Figure 6:
Graphical representation of a Colvars configuration.
The colvar called ``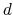 '' is defined as the difference between two distances: the first distance (
'' is defined as the difference between two distances: the first distance (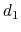 ) is taken between the center of mass of atoms 1 and 2 and that of atoms 3 to 5, the second (
) is taken between the center of mass of atoms 1 and 2 and that of atoms 3 to 5, the second (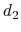 ) between atom 7 and the center of mass of atoms 8 to 10.
The difference
) between atom 7 and the center of mass of atoms 8 to 10.
The difference
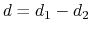 is obtained by multiplying the two by a coefficient
is obtained by multiplying the two by a coefficient 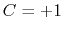 or
or 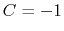 , respectively.
The colvar called ``
, respectively.
The colvar called ``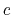 '' is the coordination number calculated between atoms 1 to 10 and atoms 11 to 20. A harmonic restraint is applied to both
'' is the coordination number calculated between atoms 1 to 10 and atoms 11 to 20. A harmonic restraint is applied to both 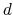 and
and 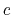 : to allow using the same force constant
: to allow using the same force constant 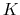 , both
, both 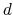 and
and 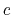 are scaled by their respective fluctuation widths
are scaled by their respective fluctuation widths 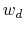 and
and 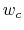 .
A third colvar ``alpha'' is defined as the
.
A third colvar ``alpha'' is defined as the 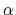 -helical content of residues 1 to 10.
The values of ``
-helical content of residues 1 to 10.
The values of ``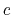 '' and ``alpha'' are also recorded throughout the simulation as a joint 2-dimensional histogram.
'' and ``alpha'' are also recorded throughout the simulation as a joint 2-dimensional histogram.
|
|
colvar {
# difference of two distances
name d
width 0.2 # 0.2 Å of estimated fluctuation width
distance {
componentCoeff 1.0
group1 { atomNumbers 1 2 }
group2 { atomNumbers 3 4 5 }
}
distance {
componentCoeff -1.0
group1 { atomNumbers 7 }
group2 { atomNumbers 8 9 10 }
}
}
colvar {
name c
coordNum {
cutoff 6.0
group1 { atomNumbersRange 1-10 }
group2 { atomNumbersRange 11-20 }
}
}
colvar {
name alpha
alpha {
psfSegID PROT
residueRange 1-10
}
}
harmonic {
colvars d c
centers 3.0 4.0
forceConstant 5.0
}
histogram {
colvars c alpha
}
Section ![[*]](crossref.png) explains how to define a colvar and its behavior, regardless of its specific functional form.
To define colvars that are appropriate to a specific physical system, Section
explains how to define a colvar and its behavior, regardless of its specific functional form.
To define colvars that are appropriate to a specific physical system, Section ![[*]](crossref.png) documents how to select atoms, and section
documents how to select atoms, and section ![[*]](crossref.png) lists all of the available functional forms, which we call ``colvar components''.
Finally, section
lists all of the available functional forms, which we call ``colvar components''.
Finally, section ![[*]](crossref.png) lists the available methods and algorithms to perform biased simulations and multidimensional analysis of colvars.
lists the available methods and algorithms to perform biased simulations and multidimensional analysis of colvars.
Aside from the Colvars configuration, an optional input state file may be provided to load the relevant data from a previous simulation.
The name of this file is provided as a value to the keyword colvarsInput.
This file contains information about the current run, including the number of the last computed step. Within Colvars, this number will be carried over to the new simulation, unless firstTimestep is given: in this case, the value of firstTimestep will always be used as first step by Colvars as well.
During a simulation with collective variables defined, the following three output files are written:
- a state file, named outputName.colvars.state; this file is in ASCII (plain text) format, regardless of the value of binaryOutput in the NAMD configuration, and is meant to be used as input when continuing a simulation (see
![[*]](crossref.png) ).
).
- if the NAMD parameter restartFreq or the parameter colvarsRestartFrequency is larger than zero, a restart file named restartName.colvars.state is written every that many steps: this file is equivalent to the final state file;
- if the parameter colvarsTrajFrequency is greater than 0 (default: 100), a trajectory file is written during the simulation: its name is outputName.colvars.traj; unlike the state file, it is not needed to restart a simulation, but can be used later for post-processing and analysis.
Other output files may be written by specific methods applied to the colvars (e.g. by the ABF method, see ![[*]](crossref.png) , or the metadynamics method, see
, or the metadynamics method, see ![[*]](crossref.png) ).
Like the colvar trajectory file, they are needed only for analyzing, not continuing a simulation.
All such files' names also begin with the prefix outputName.
).
Like the colvar trajectory file, they are needed only for analyzing, not continuing a simulation.
All such files' names also begin with the prefix outputName.
Finally, the total energy of all biases or restraints applied to the colvars appears under the NAMD standard output, under the MISC column.




Next: Defining and setting up
Up: Collective Variable-based Calculations (Colvars)1
Previous: A crash course
Contents
Index
http://www.ks.uiuc.edu/Research/namd/
![[*]](crossref.png) and in the following sections.
and in the following sections.
![[*]](crossref.png) and in the following sections.
and in the following sections.
![]() molid
molid![]() to set up the Colvars module for a given molecule.
In all following uses, the cv command will continue operating on the same molecule, regardless of its ``top'' status.
To use the cv command on a different molecule, use cv delete first and then cv molid
to set up the Colvars module for a given molecule.
In all following uses, the cv command will continue operating on the same molecule, regardless of its ``top'' status.
To use the cv command on a different molecule, use cv delete first and then cv molid ![]() molid
molid![]() .
Invoking the cv command with no arguments prints a help screen.
.
Invoking the cv command with no arguments prints a help screen.
![]() subcommand
subcommand![]() [args...].
For example, to query the value of a collective variable named myVar,
use the following syntax: set value [cv colvar myVar value].
All subcommands of cv are documented below.
[args...].
For example, to query the value of a collective variable named myVar,
use the following syntax: set value [cv colvar myVar value].
All subcommands of cv are documented below.
![[*]](crossref.png) );
);
![[*]](crossref.png) .
.
![[*]](crossref.png) for details.
for details.
![[*]](crossref.png) and
and ![[*]](crossref.png) , those within the harmonic and histogram blocks in
, those within the harmonic and histogram blocks in ![[*]](crossref.png) .
Note: except colvar, none of the keywords shown is mandatory.
.
Note: except colvar, none of the keywords shown is mandatory.
![\includegraphics[width=12cm]{figures/colvars_diagram}](img271.png)
![[*]](crossref.png) explains how to define a colvar and its behavior, regardless of its specific functional form.
To define colvars that are appropriate to a specific physical system, Section
explains how to define a colvar and its behavior, regardless of its specific functional form.
To define colvars that are appropriate to a specific physical system, Section ![[*]](crossref.png) documents how to select atoms, and section
documents how to select atoms, and section ![[*]](crossref.png) lists all of the available functional forms, which we call ``colvar components''.
Finally, section
lists all of the available functional forms, which we call ``colvar components''.
Finally, section ![[*]](crossref.png) lists the available methods and algorithms to perform biased simulations and multidimensional analysis of colvars.
lists the available methods and algorithms to perform biased simulations and multidimensional analysis of colvars.
![[*]](crossref.png) ).
).
![[*]](crossref.png) , or the metadynamics method, see
, or the metadynamics method, see ![[*]](crossref.png) ).
Like the colvar trajectory file, they are needed only for analyzing, not continuing a simulation.
All such files' names also begin with the prefix outputName.
).
Like the colvar trajectory file, they are needed only for analyzing, not continuing a simulation.
All such files' names also begin with the prefix outputName.