Rigid body docking to the Rpn11 density
Copy the refined Rpn11 yeast model from the last refinement step to the previously created dock_density folder and name it rpn11_yeast_midres.pdb.
As well, copy the rpn11_yeast_3jck_2_6479_density.mrc density file to this folder and navigate to it.
Run the following command in the terminal:
colores rpn11_yeast_3jck_2_6479_density.mrc rpn11_yeast_midres.pdb -res 3.9 -nprocs 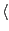 cores
cores
Adjust 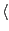 cores
cores to the number of cores you want to run Situs on.
to the number of cores you want to run Situs on.
Rename the output col_best_001.pdb to rpn11_yeast_midres_docked_3jck.pdb.
