

Next: Folding protein termini using
Up: Rosetta/MDFF Tutorial
Previous: Introduction
Contents
Subsections
Download the ModelMaker folder, which contains the files for the ModelMaker plugin to VMD. As ModelMaker wraps a multitude of different programs for computational biology, there are several
dependencies:
- VMD: Visual Molecular Dynamics; is the main program we are going to use throughout the tutorial.
It will act as the central branch point between all the other tools as well as for
visualization and analysis.
Download link:
http://www.ks.uiuc.edu/
- NAMD: Nanoscale Molecular Dynamics; is the calculation program for all Molecular Dynamics simulations we
we will carry out.
http://www.ks.uiuc.edu/
- Rosetta: is the de-novo structure prediction and refinement tool we integrated in the ModelMaker
tool. For build instructions, read the following subsection.
- Gnuplot: will be used as automated plotting program. Please install it with the package manager of your
Linux distribution or any package manager on Mac OS X.
- MODELLER: will be used for building homology models in the tutorial. Download it from
https://salilab.org/modeller/
and follow the given
installation and license request instructions.
- Situs: is a program package designed for modeling atomic structures with EM densities, such as rigid body
docking. See the download site http://situs.biomachina.org/
for download and build documentation.
Download Rosetta from https://www.rosettacommons.org. An academic license can be acquired on the homepage for free.
Download the package for your Operating System (OS). The weekly release contains the up to date software and requires about 5GB of disc space.
- 1
- Unpack the tar.zip file:
tar -xvf ROSETTA_filename.tar
- 2
- The SCons tool is used to build Rosetta alternatively to the Make build tool.
It is written in python and allows a simple multi-platform build process.
| cd $PATH_TO_ROSETTA/main/source |
|
| ./scons.py -j<number_of_processors> mode=release bin |
|
The previous command builds all executables in the release and will activate optimization flags to increase the performance.
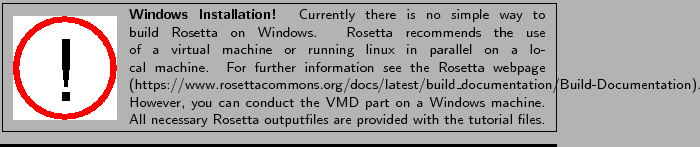
Hint: Check the OS of the workstation or cluster you want to use to run Rosetta. It is recommended to build Rosetta specifically for the target OS. If not, compatibility issues can occur during the run time. Also, make sure that build and OS versions match.
Go to the MODELLER website (https://salilab.org/modeller/tutorial/), download the newest version of MODELLER and request a license. Install MODELLER on your workstation.



Next: Folding protein termini using
Up: Rosetta/MDFF Tutorial
Previous: Introduction
Contents
www.ks.uiuc.edu/Training/Tutorials