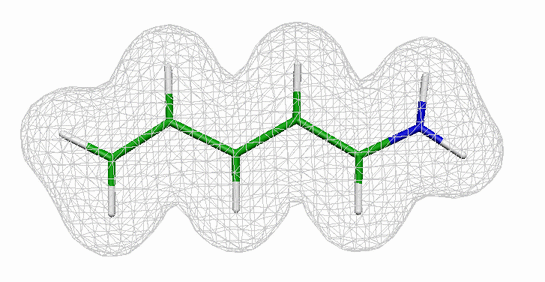
The model system that we will use to introduce you
to quantum mechanical (QM) simulations is shown in the figure on the right
(the gray wire mesh surface indicates the electron density).
The molecule is a retinal analogue and consists of one nitrogen, five carbon and
eight hydrogen atoms, which are connected to each other in a
conjugated polyene chain. The actual retinal molecule has a longer
chain and is covalently bound to a lysine residue of its host protein
via a Schiff base linkage.
Retinal is
the light absorbing moiety in the protein rhodopsin, which is responsible
for color vision in the retina of our eyes.
Before you can start studying the properties of the
retinal analogue system you need to have a well minimized
conformation of your system. You might e.g. be interested in the charge
distribution and the equilibrium bond lengths.
Since the optimization calculation can take up to 10 minutes you will first
start the minimization run and use the meantime to acquaint yourself
with the system. All the files you need for part 1 are in the directory
QM_tutorial/part1/. Go there now
All paths in the remainder of part 1 will be relative to
this directory! To start your minimization cd into the directory
min631G.
The simulation will start running and your terminal
will become inactive until the computation is finished. The simulation should
be finished in less then 15 minutes. Make sure to check, in case the shell is still
unresponsive after this amount of time has passed. In your second terminal
window you should cd into the same directory where you started the GAMESS
run and use the command
in a directory that already contains the files
filename.log, filename.dat or filename.F* (e.g. from a previous run)
it will terminate. You have to either remove or rename all files besides
filename.inp to submit a new simulation in this particular directory.
You can remove files with the rm filename command.
Since the minimization will take a few minutes
let us have a look at the retinal analogue molecule. You will not use
VMD for this
purpose, but rather the quantum chemistry visualization tool
MOLDEN
to look at GAMESS output.
Note that MOLDEN will "hijack" the terminal in which
it was started, until you terminate your MOLDEN session.
After typing the command you should see two windows: The main MOLDEN
window and the control panel shown below.
Once you feel comfortable with
MOLDEN have a closer look at your system. After you loaded the pdb file
of the retinal analogue into MOLDEN
your main window will show something like below (yours may look different
depending on your Draw Style and they way you rotated the molecule!)
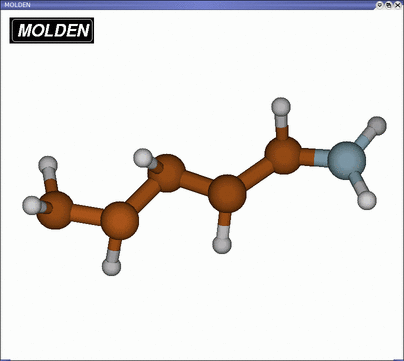
The retinal analogue consists of 5 carbon, 1 nitrogen and 8
hydrogen atoms. To practice your understanding of internal coordinates you
can use MOLDEN’s geometry tools to confirm the Z-matrix input
of the $DATA field as discussed in a previous section. Hence measure
some of the bonds, angles and dihedrals and compare with the values given
in the input file.
Note that the starting conformation of the retinal analogue is
non-planar.
Now quit MOLDEN by hitting the SKULL button in the lower center of
the control panel. Then go back into the main directory by typing
Once your simulation has finished, the
shell in which you started the job will become responsive again.
Before you do anything else you have to make sure that
your simulation terminated normally and everything went
smooth (it is a very common mistake to skip this step
and take an improperly converged result as final result; always check your
log file thoroughly!).
First check your log file for successful job completion (you can use
any editor of your choice; however, you need to know how to use it to
search for strings, since you have to go through the rather lengthy log files.
In nedit you can search with the Search -> Find menu item.
Xemacs also has a simple interface to search a file in case you don’t know how this
feature works in your editor: You can either go to Edit -> Search
or press CTRL S and then enter the string you are looking for at the
bottom of the window).
The last couple of lines should look similar to
EXECUTION OF GAMESS TERMINATED NORMALLY HH:MM:SS DD-MMM-19YY
DATA SERVER STATS: TOTAL DISTRIBUTED MEMORY USED (MEMDDI)= 0 MWORDS.
FIRST DATA SERVER’S MAXIMUM MEMORY= 0 WORDS, CPU= 0.0
SECONDS.
ddikick: all processes have ended gracefully.
unset echo
----- accounting info -----
Wed May 21 17:43:37 CDT 2003
Files used on the master node dakar.ks.uiuc.edu were:
ESC[00m-rw-r--r-- 1 markus tbres 257429 May 21 17:43 ESC[00m.//rethess.datESC[00m
-rw-r--r-- 1 markus tbres 1522 May 21 15:39 ESC[00m.//rethess.F05ESC[00m
-rw-r--r-- 1 markus tbres 1456752 May 21 17:43 ESC[00m.//rethess.F10ESC[00m
-rw-r--r-- 1 markus tbres 3100768 May 21 17:41 ESC[00m.//rethess.F22ESC[00m
-rw-r--r-- 1 markus tbres 1522 May 21 15:39 ESC[00m.//rethess.inpESC[00m
-rw-r--r-- 1 markus tbres 29985 May 21 17:43 ESC[00m.//rethess.ircESC[00m
-rw-r--r-- 1 markus tbres 562043 May 21 17:43 ESC[00m.//rethess.logESC[00m
ESC[m7101.060u 29.011s 2:04:02.62 95.8% 0+0k 0+0io 5617pf+0w
Most important is the statement
EXECUTION OF GAMESS TERMINATED
NORMALLY telling you that everything went fine (at least from the
computational point of view, this does not imply that your simulation actually
accomplished what you intended it to do).
If your simulations did not terminate with this message you have to figure out
what went wrong (contact your lab administrator if you are not sure what to do).
Since you performed a geometry optimization you also have to check if GAMESS
was able to find the equilibrium geometry or if, for some reason, this was
not the case. In the log file look for the line
1 ***** EQUILIBRIUM GEOMETRY LOCATED *****
telling you that the geometry optimization was successful.
If your simulation was not able to converge to the equilibrium geometry, try
to figure out why. Maybe the maximum number of optimization steps requested
via NSTEP in the input file was not sufficiently large.
After you convinced yourself that your simulation terminated properly
your working directory should contain the files retmin.inp, ret_analogue.log
and retmin.dat
We can now use MOLDEN to view the output
generated by GAMESS during minimization. Make sure that you are in
the directory min631G where you performed your
minimization. Start up MOLDEN
(click here if you are not sure how) and load the
log file. MOLDEN allows you to view the sequence of geometries computed
during minimization until the minimum was reached; use the three
buttons in the Select Point menu: FIRST brings you to the first
frame, NEXT to next one and MOVIE plays the whole trajectory.
Look at your output and observe the changes in structure as the system
approaches its minimal energy conformation. What is the major change and how
could you explain this behavior. Try to identify which bonds in the system
are double and which are single bonds. Any pattern?
Now that you have obtained the optimized structure
of the retinal analogue you can use GAMESS to determine some of the molecular
properties you might be interested in.
Let us assume that you would like to perform an MD simulation
of rhodopsin, which contains retinal in its binding pocket. As you know by now,
in order to do so you need to have the force field parameters of retinal,
e.g. partial charges, bond lengths, force constants etc..
These are not provided in the standard CHARMM parameter files and you
therefore may have to calculate them yourself.
The optimized structure of the retinal analogue already provides
the equilibrium values for bond lengths, angles and dihedrals as well as values
for charges as you will see shortly, but you are still missing all the
force constants.
During this session you will only consider how to obtain the force constants
for the bonds and the partial charges of the atoms.
The other parameters like the angle bending terms
can also be calculated, but are more laborious.
(You may want to use one of your evening labs to work on them.
How would you e.g. go about calculating the stiffness of the angles? You may
also want to explore how inclusion of electron correlation will
affect the results.)
In order to obtain the force constants
for the bonds you will have to calculate the Hessian matrix of
the retinal analogue in its minimized conformation.
This matrix consists of the second derivatives of the potential
energy with respect to the coordinates. Its eigenvectors are the normal modes
of the molecule. The normal modes can be projected onto the bonds of the
molecule to extract their respective vibration frequencies f and force
constants k, which are related via
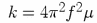 ,
where
,
where 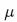 is
the reduced mass (i.e.
is
the reduced mass (i.e.
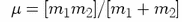 )
of the bonded atoms.
)
of the bonded atoms.
Hence, you will have to set up the run for the
calculation of the hessian matrix. Please go into the directory
hess631G to set up this run and perform your calculations.
To obtain the input file for the calculation you can start from the input file
you used for the minimization by copying it to a new file
rethess.inp.
tbss> cd ../hess631G
tbss> cp ../min631G/retmin.inp ./rethess.inp
Open the new file rethess.inp
in a text editor and delete the old internal coordinates in the
$DATA section
(make sure to only delete the coordinates, not the first two non-coordinate
lines in the file!). Next you have to insert the new, optimized internal
coordinates. They can be extracted from the log file of your minimization run
(remember: this file is located in the directory min631G).
Hence load retmin.log into your favorite editor and search for the
line
1 ***** EQUILIBRIUM GEOMETRY LOCATED *****
in the last third of the output file.
Then scroll down a little more until you see the line
THE CURRENT FULLY SUBSTITUTED Z-MATRIX IS.
The next 13 lines represent your optimized Z-matrix.
Now copy and paste the lines into your new input file
rethess.inp.
Finally you have to tell GAMESS that you would like to calculate the Hessian
matrix and that you request the projection of the force constants onto the bonds.
The first can accomplished by changing the runtyp to RUNTYP=HESSIAN
in the $CONTRL group. The projection can be requested by inserting the
keyword fields
$FORCE PURIFY=.TRUE. PRTIFC=.TRUE. DECOMP=.TRUE. $END
$ZMAT IZMAT(1)=1,1,2, 1,2,3, 1,2,4, 1,4,5, 1,5,6, 1,6,7,
1,7,8, 1,4,9, 1,5,10, 1,6,11, 1,7,12, 1,8,13,
1,8,14,
2,1,2,3, 2,3,2,4, 2,3,4,5, 2,4,5,6, 2,5,6,7,
2,6,7,8, 2,3,4,9, 2,4,5,10, 2,5,6,11,
2,6,7,12, 2,7,8,13, 2,7,8,14,
3,1,3,2,4, 3,2,3,4,5, 3,3,4,5,6, 3,4,5,6,7,
3,5,6,7,8, 3,2,3,4,9, 3,9,4,5,10,
3,10,5,6,11, 3,11,6,7,12, 3,12,7,8,13
3,12,7,8,14
$END
at the end of the input file.
Don’t forget about inserting
a single space in front of the $.
Once you are done you may compare your file with the templates in the
sample directory. Make sure that you have no
additional empty lines, tabs etc., otherwise GAMESS might get confused.
Now you are ready to submit your simulation to calculate the Hessian matrix
of the optimized retinal analogue. In the directory hess631G
type
tbss> rungms rethess.inp >& rethess.log
While the simulation is running you should
start extracting the atomic charges from the log file of your minimization run.
What you need to know are the partial atomic charges of each of the atoms in
the system. Quantum mechanics, however, only provides the electronic
wavefunction and has no concept of assigning electrons to particular atoms.
There is, therefore, no unique way of assigning partial charges with the
help of QM calculations and several schemes exist
to achieve this task. We will compare the results of three of them,
namely Mulliken, Lowdin and ESP charges.
The latter are obtained by fitting charges to the electrostatic potential,
whereas the first two follow from projecting the electron density onto
the nuclei.
To extract the Mulliken and Lowdin charges from the log file of your minimization
run, open retmin.log in a text editor.
Since these charges are calculated at each step of the minimization run you
have to make sure to pick the ones for the final minimization step.
Hence start to search from the end of the log file and look for
TOTAL MULLIKEN AND LOWDIN ATOMIC POPULATIONS
Right underneath this line you will find two columns
containing the Mulliken and Lowdin charges.
If you are not sure that you found the correct entry you can peek
here. Make sure to pick the charges and not
the populations. The ESP charges you will find almost at the end of the log file
below the line titled
(check here if you are not sure).
Now you have three sets of partial atomic charges for your system.
You should compare them and notice that they are similar, but far from being
identical.
Partical charges for force fields are typically determined
using so called restrained ESP charges at a HF/6-31G* level of theory,
which are closely related to the ESP charges calculated by us. The reason
being the fact that they are much less sensitive to a change in basis
than the Mulliken or Lowdin charges. You can convince yourself that
this is the case by using one of the evening sessions (don’t do it
now) to repeat the charge calculation using the HF/6-31+G level of theory,
which includes diffuse functions (consult the manual to find the correct keyword.
HINT: look in the BASIS section).
You may also compare your computed set of partial charges with CHARMM values
which were used in an actual MD simulation
(CHARMM charges for retinal analogue).
Once the calculation of your Hessian matrix
has finished have a look at the results.
Make sure, as in the case of the minimization, that your
GAMESS run has executed normally and nothing unexpected has happened.
(note that this time you do not need to check for 1 ***** EQUILIBRIUM
GEOMETRY LOCATED ***** since you did not perform an optimization, but
a calculation of the Hessian matrix!)
Use MOLDEN to look at the output file
rethess.log. Hence start up MOLDEN, load the file and
adjust the view of the molecule to your liking. Then press the Norm Mode
button and a window will pop up that will allow you to view the normal modes of the
retinal analogue that you have just calculated. The modes are ordered according to
increasing frequencies (i.e. energy) and you can view them by simply
clicking on them.
You should try to answer the following questions: Why do the first 6 normal modes
have zero frequency and what are they? Can you discern qualitatively what
distinguishes the low frequency modes (i.e. low energy)
from the high frequency ones (i.e. high energy)?
In order to extract the force constants for the bonds
which follow from the hessian matrix you just calculated, have a look at
the log file rethess.log. Hence, load the file into an
editor and look for the lines
--------------------------------------------------------------------
INTERNAL COORDINATE INTRINSIC FORCE CONSTANTS
(HARTREES/BOHR**2) (MDYN/ANG)
--------------------------------------------------------------------
The table right underneath lists the force constants for the
13 bonds in the retinal analogue in the same order as in the Z-matrix
in the $DATA section of the input file (note that e.g. STR. 1 2
corresponds to the bond between atom 1 and 2).
The figure below gives the atom numbering
.
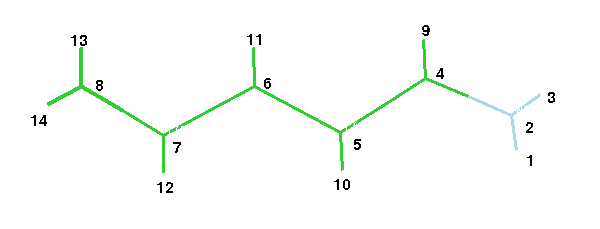
They are given in
Hartree/Bohr2 and you should convert them into
kcal/(molÅ)2) to be able to use them in a CHARMM
parameter file (note: 1 Hartree = 627.53 kcal/mol,
1 Bohr = 0.529Å).
Your calculation of bond frequencies/force constants
is based on an harmonic approximation for the potential (remember, the
Hessian is the second derivative of the potential energy function!).
This generally overestimates the vibration frequencies and
workers in the field often correct this by multiplying the frequencies f with
by factor of 0.9 (what do you need to multiply the force constants with,
considering that 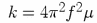 ?)
?)
Lastly, and quite unfortunately, the CHARMM force constants
are defined via 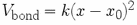 , therefore
lacking the conventional factor of one half.
Hence, you also have to multiply your force constants by a
factor of 0.5 before you can use them in a CHARMM parameter file.
You can either do the calculations by hand using a calculator, or
copy and paste the 13 lines with the force constants into a text editor
and save them to the file force.dat. You can then use
the command
, therefore
lacking the conventional factor of one half.
Hence, you also have to multiply your force constants by a
factor of 0.5 before you can use them in a CHARMM parameter file.
You can either do the calculations by hand using a calculator, or
copy and paste the 13 lines with the force constants into a text editor
and save them to the file force.dat. You can then use
the command
tbss> gawk '{print $4*0.5*0.81*627.53/(0.529^2)}' force.dat > force_final.dat
to obtain the file force_final.dat with the
converted force constants. Use the above figure of the retinal
analogue to assign the force constants
to the respective bonds and best write them into the figure. Next use MOLDEN to
measure the bond distances and also enter them into the figure. Finally,
put down the ESP charges of each atom. You have now collected all the
information you need to complete the topology and parameter files for the
retinal analogue. There is one last thing that you need to consider.
In the actual parameter file the bonds between atoms 8-13 and 8-14
have to be the same due to symmetry (the same is true for the bonds between
1-2 and 2-3!). There will hence only be one entry for each type of bond
(this is why in the parameter file there are only eleven bond entries
to complete and not 13). Since you probably have different values for
the force constants and lengths of the bonds 8-13 and 8-14 just
take the average! Finally, go back into the main directory
Finally, after you put all that effort into calculating
the minimum energy conformation, ESP partial atomic charges, bond lengths,
angles etc. you can now put them to use and complete the CHARMM
parameter and topology files for our retinal analogue.
After generating the psf file using psfgen you can then use
NAMD to classically minimize the original conformation of the retinal analogue
using your newly designed force field.
Hence go into the directory
namdmin/toppar
which contains the parameter and topology files for the retinal analogue (the
values you need to fill in are denoted with Xs).
Then fill in your calculated ESP charges
(they go into the topology file as you know by now; note that the indices of
the atoms in the topology file differs slightly from the ones in the
Z-matrix and the above
figure should help you to identify which correspond to which), the bond
lengths and finally the force constants for the bonds that you have calculated
(they go into the parameter file! If you have doubts about the correctness of
your newly designed files you may peek at at the provided files in the
sample directory. If your values differ slightly,
that’s all right!)
You don’t have to worry about the angle and dihedral terms: leave them
as they are. Once you have accomplished this, go into the directory
namdmin/psf directory
and create the psf file of the retinal analogue (remember: you need
the original pdb file and the topology file that you just created;
hence copy them into the directory).
tbss> cp ../toppar/top_ret.inp ./
tbss> cp ../../pdbfile/ret.pdb ./
Open VMD and use psfgen to generate the psf file
(you should be able to this yourself by now; in case you run into serious
trouble generating the psf file you may use the create.png
script in the sample directory). Remember: To do so you need the
completed topology file as well as the original pdb file. Please do not close
VMD since we will need it again in a minute.
You are now ready to employ NAMD to minimize the retinal analogue structure
using your own parameter set (1000 minimization steps are more than sufficient).
Please do this in the directory
namdmin/min, hence
We encourage you to set up the NAMD input file yourself, but you may also use
the provided one in the sample directory. You also need to copy
the pdb and psf files from the create_psf directory as well
as the completed parameter file from the directory toppar
into this directory. Then start
the simulation by typing:
tbss> namd2 [name of inputfile] >& [name of outputfile]
After your minimization has finished, have a look at your final structure
and compare with the QM optimization. How did your parameters perform?
The following provides some suggestions for further
study during your evening sessions. Please don’t do the exercises during
the tutorial, but rather go on with part 2.
We encourage you to repeat part 1 using a different, preferably
larger, basis set, e.g. HF/6-31G* or HF/6-31+G*.
In order to speed up your computations
you can start your minimization from the optimized 6-31G geometry.
Examine the changes in energy, structure and parameters (charges, bond lengths,
force constants) when using a different basis set. This will show you the importance
of carefully choosing the basis set of QM calculations in order to
obtain meaningful results. It is also important to consider the influence
of electron correlation by repeating the above calculations using density
functional theory (DFT). Since this will be much more time consuming you will have
to do this during one of the evening sessions. In order to employ DFT you
simply insert the following line into you input file
which will use the B3LYP functional to take
into account electronic correlation effects.


Next:
Up:
Previous:
markus@ks.uiuc.edu